Biostatistics
Francis Galton tried to expand Mendel's discoveries with human data and proposed a different model with fractions of the heredity coming from each ancestral composing an infinite series.
His ideas were strongly disagreed by William Bateson, who followed Mendel's conclusions, that genetic inheritance were exclusively from the parents, half from each of them.
By the 1930s, models built on statistical reasoning had helped to resolve these differences and to produce the neo-Darwinian modern evolutionary synthesis.
These and other biostatisticians, mathematical biologists, and statistically inclined geneticists helped bring together evolutionary biology and genetics into a consistent, coherent whole that could begin to be quantitatively modeled.
In parallel to this overall development, the pioneering work of D'Arcy Thompson in On Growth and Form also helped to add quantitative discipline to biological study.
Despite the fundamental importance and frequent necessity of statistical reasoning, there may nonetheless have been a tendency among biologists to distrust or deprecate results which are not qualitatively apparent.
One anecdote describes Thomas Hunt Morgan banning the Friden calculator from his department at Caltech, saying "Well, I am like a guy who is prospecting for gold along the banks of the Sacramento River in 1849.
The correct definition of the main hypothesis and the research plan will reduce errors while taking a decision in understanding a phenomenon.
It is essential to carry the study based on the three basic principles of experimental statistics: randomization, replication, and local control.
The research will be headed by the question, so it needs to be concise, at the same time it is focused on interesting and novel topics that may improve science and knowledge and that field.
The main propose is called null hypothesis (H0) and is usually based on a permanent knowledge about the topic or an obvious occurrence of the phenomena, sustained by a deep literature review.
In clinical research, the trial type, as inferiority, equivalence, and superiority is a key in determining sample size.
It is common to use randomized controlled clinical trials, where results are usually compared with observational study designs such as case–control or cohort.
[6] Data collection methods must be considered in research planning, because it highly influences the sample size and experimental design.
For qualitative data, collection can be done with structured questionnaires or by observation, considering presence or intensity of disease, using score criterion to categorize levels of occurrence.
Especially, in genetic studies, modern methods for data collection in field and laboratory should be considered, as high-throughput platforms for phenotyping and genotyping.
These tools allow bigger experiments, while turn possible evaluate many plots in lower time than a human-based only method for data collection.
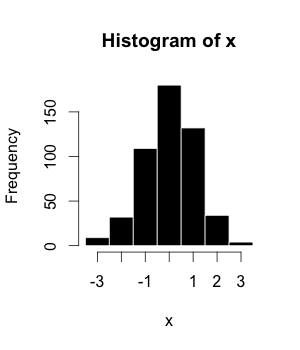
The histogram (or frequency distribution) is a graphical representation of a dataset tabulated and divided into uniform or non-uniform classes.
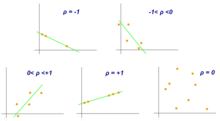
Although correlations between two different kinds of data could be inferred by graphs, such as scatter plot, it is necessary validate this though numerical information.
In other words, it is desirable to obtain parameters to describe the population of interest, but since the data is limited, it is necessary to make use of a representative sample in order to estimate them.
[18] Verifying whether the outcome of a statistical test does not change when the technical assumptions are slightly altered (so-called robustness checks) is the main way of combating mis-specification.
New biomedical technologies like microarrays, next-generation sequencers (for genomics) and mass spectrometry (for proteomics) generate enormous amounts of data, allowing many tests to be performed simultaneously.
They are useful for researchers depositing data, retrieve information and files (raw or processed) originated from other experiments or indexing scientific articles, as PubMed.
[23] Phytozome,[24] in turn, stores the assemblies and annotation files of dozen of plant genomes, also containing visualization and analysis tools.
[28] Nowadays, increase in size and complexity of molecular datasets leads to use of powerful statistical methods provided by computer science algorithms which are developed by machine learning area.
Collaborative work among molecular biologists, bioinformaticians, statisticians and computer scientists is important to perform an experiment correctly, going from planning, passing through data generation and analysis, and ending with biological interpretation of the results.
[30] However, QTL mapping resolution is impaired by the amount of recombination assayed, a problem for species in which it is difficult to obtain large offspring.
While QTL mapping is limited due resolution, GWAS does not have enough power when rare variants of small effect that are also influenced by environment.
As a summary, some points about the application of quantitative genetics are: Studies for differential expression of genes from RNA-Seq data, as for RT-qPCR and microarrays, demands comparison of conditions.
In RNA-Seq, the quantification of expression uses the information of mapped reads that are summarized in some genetic unit, as exons that are part of a gene sequence.