Carl Woese
Woese is famous for defining the Archaea (a new domain of life) in 1977 through a pioneering phylogenetic taxonomy of 16S ribosomal RNA, a technique that has revolutionized microbiology.
[12] In 1953, he completed a PhD in biophysics at Yale University, where his doctoral research focused on the inactivation of viruses by heat and ionizing radiation.
[13][16] In 1964, Woese joined the microbiology faculty of the University of Illinois Urbana–Champaign, where he focused on Archaea, genomics, and molecular evolution as his areas of expertise.
[16] Woese died on December 30, 2012, following complications from pancreatic cancer, leaving as survivors his wife Gabriella and a son and daughter.
[20] He then re-evaluated experimental data associated with the hypothesis that viruses used one base, rather than a triplet, to encode each amino acid, and suggested 18 codons, correctly predicting one for proline.
[14][21] Other work established the mechanistic basis of protein translation, but in Woese's view, largely overlooked the genetic code's evolutionary origins as an afterthought.
[14] With the freedom to patiently pursue more speculative threads of inquiry outside the mainstream of biological research, Woese began to consider the genetic code in evolutionary terms, asking how the codon assignments and their translation into an amino acid sequence might have evolved.
[23][24] Adapted from Édouard Chatton's generalization, Stanier and Van Niel's concept was quickly accepted as the most important distinction among organisms; yet they were nevertheless skeptical of microbiologists' attempts to construct a natural phylogenetic classification of bacteria.
[25] However, it became generally assumed that all life shared a common prokaryotic (implied by the Greek root πρό (pro-), before, in front of) ancestor.
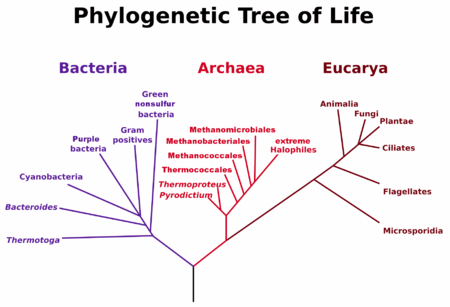
[24][26] In 1977, Woese and George E. Fox experimentally disproved this universally held hypothesis about the basic structure of the tree of life.
His three-domain system, based on phylogenetic relationships rather than obvious morphological similarities, divided life into 23 main divisions, incorporated within three domains: Bacteria, Archaea, and Eucarya.
A decade of labor-intensive oligonucleotide cataloging left him with a reputation as "a crank," and Woese would go on to be dubbed as "Microbiology's Scarred Revolutionary" by a news article printed in the journal Science.
[30] Organisms similar to those archaea that exist in extreme environments may have developed on other planets, some of which harbor conditions conducive to extremophile life.
Using ribosomal RNA sequence as an evolutionary measure, my laboratory has reconstructed the phylogeny of both groups, and thereby provided a phylogenetically valid system of classification for prokaryotes.
[27][33] First described by Woese and Fox in a 1977 paper and explored further with microbiologist Jane Gibson in a 1980 paper, these organisms, or progenotes, were imagined as protocells with very low complexity due to their error-prone translation apparatus ("noisy genetic transmission channel"), which produced high mutation rates that limited the specificity of cellular interaction and the size of the genome.
[27] Furthermore, because of this reduced specificity, all cellular components were susceptible to horizontal gene transfer, and rapid evolution occurred at the level of the ecosystem.
[33] In later years, Woese's work concentrated on genomic analysis to elucidate the significance of horizontal gene transfer (HGT) for evolution.
[37] He worked on detailed analyses of the phylogenies of the aminoacyl-tRNA synthetases and on the effect of horizontal gene transfer on the distribution of those key enzymes among organisms.
[38] The goal of the research was to explain how the primary cell types (the archaeal, eubacterial, and eukaryotic) evolved from an ancestral state in the RNA world.