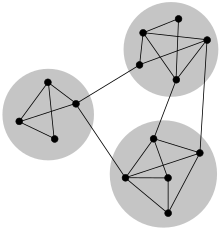
Community structure
In the particular case of non-overlapping community finding, this implies that the network divides naturally into groups of nodes with dense connections internally and sparser connections between groups.
Social networks include community groups (the origin of the term, in fact) based on common location, interests, occupation, etc.
[5][6] Finding an underlying community structure in a network, if it exists, is important for a number of reasons.
In metabolic networks, such functional groups correspond to cycles or pathways whereas in the protein interaction network, communities correspond to proteins with similar functionality inside a biological cell.
Thus, only concentrating on the average properties usually misses many important and interesting features inside the networks.
For example, in a given social network, both gregarious and reticent groups might exists simultaneously.
Both these cases are well handled by community detection algorithm since it allows one to assign the probability of existence of an edge between a given pair of nodes.
Despite these difficulties, however, several methods for community finding have been developed and employed with varying levels of success.
This method sees use, for example, in load balancing for parallel computing in order to minimize communication between processor nodes.
[10] Another method for finding community structures in networks is hierarchical clustering.
Commonly used measures include the cosine similarity, the Jaccard index, and the Hamming distance between rows of the adjacency matrix.
An important step is how to determine the threshold to stop the agglomerative clustering, indicating a near-to-optimal community structure.
A common strategy consist to build one or several metrics monitoring global properties of the network, which peak at given step of the clustering.
An interesting approach in this direction is the use of various similarity or dissimilarity measures, combined through convex sums,.
[11] Another approximation is the computation of a quantity monitoring the density of edges within clusters with respect to the density between clusters, such as the partition density, which has been proposed when the similarity metric is defined between edges (which permits the definition of overlapping communities),[12] and extended when the similarity is defined between nodes, which allows to consider alternative definitions of communities such as guilds (i.e. groups of nodes sharing a similar number of links with respect to the same neighbours but not necessarily connected themselves).
The Girvan–Newman algorithm returns results of reasonable quality and is popular because it has been implemented in a number of standard software packages.
[14] In spite of its known drawbacks, one of the most widely used methods for community detection is modularity maximization.
[14] Modularity is a benefit function that measures the quality of a particular division of a network into communities.
Since exhaustive search over all possible divisions is usually intractable, practical algorithms are based on approximate optimization methods such as greedy algorithms, simulated annealing, or spectral optimization, with different approaches offering different balances between speed and accuracy.
[15][16] A popular modularity maximization approach is the Louvain method, which iteratively optimizes local communities until global modularity can no longer be improved given perturbations to the current community state.
[20] Methods based on statistical inference attempt to fit a generative model to the network data, which encodes the community structure.
The overall advantage of this approach compared to the alternatives is its more principled nature, and the capacity to inherently address issues of statistical significance.
Most methods in the literature are based on the stochastic block model[21] as well as variants including mixed membership,[22][23] degree-correction,[24] and hierarchical structures.
[29] Currently many algorithms exist to perform efficient inference of stochastic block models, including belief propagation[30][31] and agglomerative Monte Carlo.
The simplest is to consider only maximal cliques bigger than a minimum size (number of nodes).
For Euclidean spaces, methods like embedding-based Silhouette community detection[38] can be utilized.
Other more flexible benchmarks have been proposed that allow for varying group sizes and nontrivial degree distributions, such as LFR benchmark[41] [42] which is an extension of the four groups benchmark that includes heterogeneous distributions of node degree and community size, making it a more severe test of community detection methods.
[43][44] Commonly used computer-generated benchmarks start with a network of well-defined communities.
They compare the solution obtained by an algorithm [42] with the original community structure, evaluating the similarity of both partitions.
This transition is independent of the type of algorithm being used to detect communities, implying that there exists a fundamental limit on our ability to detect communities in networks, even with optimal Bayesian inference (i.e., regardless of our computational resources).