Computational anatomy
Computational anatomy is an interdisciplinary field of biology focused on quantitative investigation and modelling of anatomical shapes variability.
[3] It also implies that the diffeomorphic shape momentum taken pointwise satisfying the Euler–Lagrange equation for geodesics is determined by its neighbors through spatial derivatives on the velocity field.
Computational anatomy intersects the study of Riemannian manifolds and nonlinear global analysis, where groups of diffeomorphisms are the central focus.
The spirit of this discipline shares strong overlap with areas such as computer vision and kinematics of rigid bodies, where objects are studied by analysing the groups responsible for the movement in question.
The models of metric pattern theory,[12][13] in particular group action on the orbit of shapes and forms is a central tool to the formal definitions in computational anatomy.
[17][18][19] The first formalization of computational anatomy as an orbit of exemplar templates under diffeomorphism group action was in the original lecture given by Grenander and Miller with that title in May 1997 at the 50th Anniversary of the Division of Applied Mathematics at Brown University,[20] and subsequent publication.
They are generated via Lagrangian and Eulerian flows which satisfy a law of composition of functions forming the group property, but are not additive.
The Lagrangian and Hamiltonian formulations of the equations of motion of computational anatomy took off post 1997 with several pivotal meetings including the 1997 Luminy meeting[21] organized by the Azencott[22] school at Ecole-Normale Cachan on the "Mathematics of Shape Recognition" and the 1998 Trimestre at Institute Henri Poincaré organized by David Mumford "Questions Mathématiques en Traitement du Signal et de l'Image" which catalyzed the Hopkins-Brown-ENS Cachan groups and subsequent developments and connections of computational anatomy to developments in global analysis.
These contributions of computational anatomy to the global analysis associated to the infinite dimensional manifolds of subgroups of the diffeomorphism group is far from trivial.
The original idea of doing differential geometry, curvature and geodesics on infinite dimensional manifolds goes back to Bernhard Riemann's Habilitation (Ueber die Hypothesen, welche der Geometrie zu Grunde liegen[49][50]); the key modern book laying the foundations of such ideas in global analysis are from Michor.
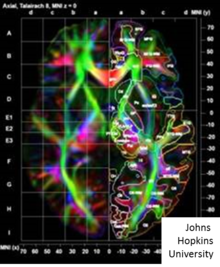
[51] The applications within medical imaging of computational anatomy continued to flourish after two organized meetings at the Institute for Pure and Applied Mathematics conferences[52][53] at University of California, Los Angeles.
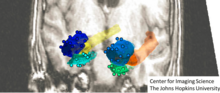
[58][59] Atlas based methods and virtual textbooks[60] which accommodate variations as in deformable templates are at the center of many neuro-image analysis platforms including Freesurfer,[61] FSL,[62] MRIStudio,[63] SPM.
[citation needed] For the study of rigid body kinematics, the low-dimensional matrix Lie groups have been the central focus.
The matrix groups are low-dimensional mappings, which are diffeomorphisms that provide one-to-one correspondences between coordinate systems, with a smooth inverse.
To ensure smooth flows of diffeomorphisms avoiding shock-like solutions for the inverse, the vector fields must be at least 1-time continuously differentiable in space.
Oftentimes, the familiar Euclidean metric is not directly applicable because the patterns of shapes and images do not form a vector space.
The Euler–Lagrange equation in computational anatomy describes the geodesic shortest path flows between coordinate systems of the diffeomorphism metric.
[83] In Riemannian Metric and Lie-Bracket Interpretation of the Euler–Lagrange Equation on Geodesics derivations are provided in terms of the adjoint operator and the Lie bracket for the group of diffeomorphisms.
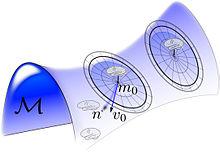
In computational anatomy the diffeomorphisms are used to push the coordinate systems, and the vector fields are used as the control within the anatomical orbit or morphological space.
For landmarks[90][91][92] the geodesics have Eulerian shape momentum which are a superposition of delta distributions travelling with the finite numbers of particles; the diffeomorphic flow of coordinates have velocities in the range of weighted Green's Kernels.
Large deformations began in the early 1990s,[18][19] with the first existence to solutions to the variational problem for flows of diffeomorphisms for dense image matching established in.
gives the variational problem Beg's iterative LDDMM algorithm has fixed points which satisfy the necessary optimizer conditions.
gives the variational problem Viallard's iterative Hamiltonian LDDMM has fixed points which satisfy the necessary optimizer conditions.
This LDDMM-ODF mapping algorithm has been widely used to study brain white matter degeneration in aging, Alzheimer's disease, and vascular dementia.
In this setting metamorphosis combines both the diffeomorphic coordinate system transformation of computational anatomy as well as the early morphing technologies which only faded or modified the photometric or image intensity alone.
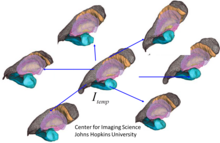
Matching geometrical objects like unlabelled point distributions, curves or surfaces is another common problem in computational anatomy.
Even in the discrete setting where these are commonly given as vertices with meshes, there are no predetermined correspondences between points as opposed to the situation of landmarks described above.
This can be achieved via concepts and methods borrowed from Geometric measure theory, in particular currents[40] and varifolds[45] which have been used extensively for curve and surface matching.
The endpoint condition with conservation implies the initial momentum at the identity of the group: The iterative algorithm for large deformation diffeomorphic metric mapping for landmarks is given.
Statistical shape in computational anatomy is the empirical study of diffeomorphic correspondences between populations and common template coordinate systems.













![{\displaystyle v_{t},t\in [0,1]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/56e95a746da6e27d7a2c9948d7626ebaa9e80893)








