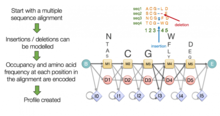
Multiple sequence alignment
A direct method for producing an MSA uses the dynamic programming technique to identify the globally optimal alignment solution.
For proteins, this method usually involves two sets of parameters: a gap penalty and a substitution matrix assigning scores or probabilities to the alignment of each possible pair of amino acids based on the similarity of the amino acids' chemical properties and the evolutionary probability of the mutation.
For nucleotide sequences, a similar gap penalty is used, but a much simpler substitution matrix, wherein only identical matches and mismatches are considered, is typical.
Expressed with the big O notation commonly used to measure computational complexity, a naïve MSA takes O(LengthNseqs) time to produce.
[5][6][7] In 1989, based on Carrillo-Lipman Algorithm,[8] Altschul introduced a practical method that uses pairwise alignments to constrain the n-dimensional search space.
[10] In 2019, Hosseininasab and van Hoeve showed that by using decision diagrams, MSA may be modeled in polynomial space complexity.
[3] The most widely used approach to multiple sequence alignments uses a heuristic search known as progressive technique (also known as the hierarchical or tree method) developed by Da-Fei Feng and Doolittle in 1987.
[13][14] ClustalW is used extensively for phylogenetic tree construction, in spite of the author's explicit warnings that unedited alignments should not be used in such studies and as input for protein structure prediction by homology modeling.
They recommend Clustal Omega which performs based on seeded guide trees and HMM profile-profile techniques for protein alignments.
Because progressive methods are heuristics that are not guaranteed to converge to a global optimum, alignment quality can be difficult to evaluate and their true biological significance can be obscure.
A semi-progressive method that improves alignment quality and does not use a lossy heuristic while running in polynomial time has been implemented in the program PSAlign.
[12] A variety of subtly different iteration methods have been implemented and made available in software packages; reviews and comparisons have been useful but generally refrain from choosing a "best" technique.
[12] Another iterative program, DIALIGN, takes an unusual approach of focusing narrowly on local alignments between sub-segments or sequence motifs without introducing a gap penalty.
An alternative method that uses fast local alignments as anchor points or seeds for a slower global-alignment procedure is implemented in the CHAOS/DIALIGN suite.
HMMs can produce a single highest-scoring output but can also generate a family of possible alignments that can then be evaluated for biological significance.
Although HMM-based methods have been developed relatively recently, they offer significant improvements in computational speed, especially for sequences that contain overlapping regions.
HHsearch[27] is a software package for the detection of remotely related protein sequences based on the pairwise comparison of HMMs.
These problems are common in newly produced sequences that are poorly annotated and may contain frame-shifts, wrong domains or non-homologous spliced exons.
A variety of methods for isolating the motifs have been developed, but all are based on identifying short highly conserved patterns within the larger alignment and constructing a matrix similar to a substitution matrix that reflects the amino acid or nucleotide composition of each position in the putative motif.
[33] Block scoring generally relies on the spacing of high-frequency characters rather than on the calculation of an explicit substitution matrix.
[34][35] Non-coding DNA regions, especially transcription factor binding sites (TFBSs), are conserved, but not necessarily evolutionarily related, and may have converged from non-common ancestors.
Standard optimization techniques in computer science – both of which were inspired by, but do not directly reproduce, physical processes – have also been used in an attempt to more efficiently produce quality MSAs.
One such technique, genetic algorithms, has been used for MSA production in an attempt to broadly simulate the hypothesized evolutionary process that gave rise to the divergence in the query set.
Example algorithms used to solve mixed integer programming models of MSA include branch and price[40] and Benders decomposition.
[3] Although exact approaches are computationally slow compared to heuristic algorithms for MSA, they are guaranteed to reach the optimal solution eventually, even for large-size problems.
In January 2017, D-Wave Systems announced that its qbsolv open-source quantum computing software had been successfully used to find a faster solution to the MSA problem.
As the number of sequence and their divergence increases many more errors will be made simply because of the heuristic nature of MSA algorithms.
[42] However, as the number of sequences increases and especially in genome-wide studies that involve many MSAs it is impossible to manually curate all alignments.
Its extension, Transitive Consistency Score (TCS), uses T-Coffee libraries of pairwise alignments to evaluate any third party MSA.
The HoT (Heads-Or-Tails) score can be used as a measure of site-specific alignment uncertainty due to the existence of multiple co-optimal solutions.