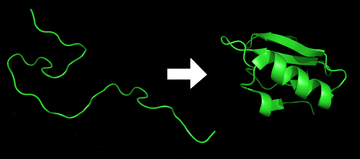
Protein folding
Several neurodegenerative and other diseases are believed to result from the accumulation of amyloid fibrils formed by misfolded proteins, the infectious varieties of which are known as prions.
[7] On the other hand, very small single-domain proteins with lengths of up to a hundred amino acids typically fold in a single step.
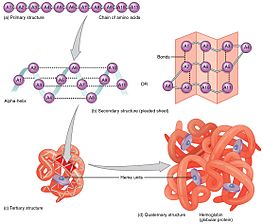
[11] The specific amino acid residues and their position in the polypeptide chain are the determining factors for which portions of the protein fold closely together and form its three-dimensional conformation.
[12] The essential fact of folding, however, remains that the amino acid sequence of each protein contains the information that specifies both the native structure and the pathway to attain that state.
[12] Folding is a spontaneous process that is mainly guided by hydrophobic interactions, formation of intramolecular hydrogen bonds, van der Waals forces, and it is opposed by conformational entropy.
[18] While these macromolecules may be regarded as "folding themselves", the process also depends on the solvent (water or lipid bilayer),[19] the concentration of salts, the pH, the temperature, the possible presence of cofactors and of molecular chaperones.
[24] In proteins with globular folds, hydrophobic amino acids tend to be interspersed along the primary sequence, rather than randomly distributed or clustered together.
[25][26] However, proteins that have recently been born de novo, which tend to be intrinsically disordered,[27][28] show the opposite pattern of hydrophobic amino acid clustering along the primary sequence.
[31] In this way, chaperones do not actually increase the rate of individual steps involved in the folding pathway toward the native structure; instead, they work by reducing possible unwanted aggregations of the polypeptide chain that might otherwise slow down the search for the proper intermediate and they provide a more efficient pathway for the polypeptide chain to assume the correct conformations.
Heat shock proteins have been found in all species examined, from bacteria to humans, suggesting that they evolved very early and have an important function.
Chaperones are shown to exist in increasing concentrations during times of cellular stress and help the proper folding of emerging proteins as well as denatured or misfolded ones.
Temperatures above or below the range that cells tend to live in will cause thermally unstable proteins to unfold or denature (this is why boiling makes an egg white turn opaque).
[40] Like GroES, gp31 forms a stable complex with GroEL chaperonin that is absolutely necessary for the folding and assembly in vivo of the bacteriophage T4 major capsid protein gp23.
[43] The structural stability of these fibrillar assemblies is caused by extensive interactions between the protein monomers, formed by backbone hydrogen bonds between their β-strands.
[42] The amyloids are fibrillary structures that contain intermolecular hydrogen bonds which are highly insoluble and made from converted protein aggregates.
The X-rays specifically interact with the electron clouds surrounding the individual atoms within the protein crystal lattice and produce a discernible diffraction pattern.
[49] Without the relation established through a mathematical basis known as Fourier transform, the "phase problem" would render predicting the diffraction patterns very difficult.
[15] Emerging methods like multiple isomorphous replacement use the presence of a heavy metal ion to diffract the X-rays into a more predictable manner, reducing the number of variables involved and resolving the phase problem.
Upon disruption of the protein's tertiary or quaternary structure, these side chains become more exposed to the hydrophilic environment of the solvent, and their quantum yields decrease, leading to low fluorescence intensities.
[52][53] General equations have been developed by Hugues Bedouelle to obtain the thermodynamic parameters that characterize the unfolding equilibria for homomeric or heteromeric proteins, up to trimers and potentially tetramers, from such profiles.
[50] Fluorescence spectroscopy can be combined with fast-mixing devices such as stopped flow, to measure protein folding kinetics,[54] generate a chevron plot and derive a Phi value analysis.
Fast techniques in use include neutron scattering,[62] ultrafast mixing of solutions, photochemical methods, and laser temperature jump spectroscopy.
Among the many scientists who have contributed to the development of these techniques are Jeremy Cook, Heinrich Roder, Terry Oas, Harry Gray, Martin Gruebele, Brian Dyer, William Eaton, Sheena Radford, Chris Dobson, Alan Fersht, Bengt Nölting and Lars Konermann.
[65] Optical tweezers have been used to stretch single protein molecules from their C- and N-termini and unfold them to allow study of the subsequent refolding.
It discovered – using single molecule optical tweezers measurement – that calcium-bound vWF acts as a shear force sensor in the blood.
Shear force leads to unfolding of the A2 domain of vWF, whose refolding rate is dramatically enhanced in the presence of calcium.
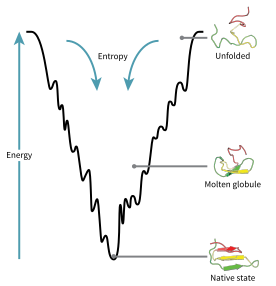
A consequence of these evolutionarily selected sequences is that proteins are generally thought to have globally "funneled energy landscapes" (a term coined by José Onuchic)[74] that are largely directed toward the native state.
[77] As the protein begins to fold and assume its various conformations, it always seeks a more thermodynamically favorable structure than before and thus continues through the energy funnel.
[80] Because of computational cost, ab initio MD folding simulations with explicit water are limited to peptides and small proteins.
Long continuous-trajectory simulations have been performed on Anton, a massively parallel supercomputer designed and built around custom ASICs and interconnects by D. E. Shaw Research.