PubMed
[2] PubMed, first released in January 1996, ushered in the era of private, free, home- and office-based MEDLINE searching.
[14] PubMed/MEDLINE can be accessed via handheld devices, using for instance the "PICO" option (for focused clinical questions) created by the NLM.
In addition, it saves the disadvantage of the free text search in which the spelling, singular/plural or abbreviated differences have to be taken into consideration.
Therefore, to guarantee an exhaustive search, a combination of controlled language headings and free text terms must be used.
Such parameters are: Article Type (MeSH terms, e.g., "Clinical Trial"), Secondary identifiers, (MeSH terms), Language, Country of the Journal or publication history (e-publication date, print journal publication date).
The secondary identifier field is to store accession numbers to various databases of molecular sequence data, gene expression or chemical compounds and clinical trial IDs.
To create these lists of related articles, PubMed compares words from the title and abstract of each citation, as well as the MeSH headings assigned, using a powerful word-weighted algorithm.
[24] This feature makes PubMed searches more sensitive and avoids false-negative (missed) hits by compensating for the diversity of medical terminology.
[27] Some 3,200 sites (mainly academic institutions) participate in this NLM facility (as of March 2010[update]), from Aalborg University in Denmark to ZymoGenetics in Seattle.
[31][32] askMEDLINE, a free-text, natural language query tool for MEDLINE/PubMed, developed by the NLM, also suitable for handhelds.
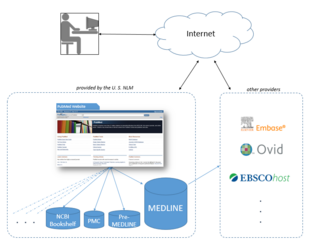
The National Library of Medicine leases the MEDLINE information to a number of private vendors such as Embase, Ovid, Dialog, EBSCO, Knowledge Finder and many other commercial, non-commercial, and academic providers.
As licenses to use MEDLINE data are available for free, the NLM in effect provides a free testing ground for a wide range[37] of alternative interfaces and 3rd party additions to PubMed, one of a very few large, professionally curated databases which offers this option.
Lu identifies a sample of 28 current and free Web-based PubMed versions, requiring no installation or registration, which are grouped into four categories:[37] As most of these and other alternatives rely essentially on PubMed/MEDLINE data leased under license from the NLM/PubMed, the term "PubMed derivatives" has been suggested.
[48] Various citation format generators, taking PMID numbers as input, are examples of web applications making use of the eutils-application program interface.
[50] Recent advancements in this field include the development of models like PubMedGPT, a 2.7B parameter model trained on PubMed data by Stanford CRFM, and Microsoft's BiomedCLIP-PubMedBERT, which utilizes figure-caption pairs from PubMed Central for vision-language processing.
These models demonstrate the significant potential of PubMed data in enhancing the capabilities of AI in medical research and healthcare applications.
Such advancements underline the growing intersection between large-scale data mining and AI development in the biomedical field.