Zoonotic origins of COVID-19
[8] By 2010, in vitro experiments had confirmed that modifications to the spike protein receptor binding domain could enable human infection by several SARS-related coronaviruses.
[9] Virologists Rachel Graham and Ralph S. Baric at that time wrote, "that the question of emergence of another pathogenic human coronavirus from bat reservoirs might be more appropriately expressed as 'when' than as 'if'.
[16] Bats, along with their viruses, have large overlapping geographic ranges in Southeast Asia,[17] and there is a particularly great concentration and diversity of bat-related coronaviruses in Southern and Southwest China.
[19] Temmam et al. found no serological evidence for exposure to BANAL-52 among bat handlers and guano collectors in the area of Laos where it was sampled.
[18] Viruses including BANAL-52 isolated from bats in Laos showed high similarity to the SARS-CoV-2 receptor binding domain in amino acid residues but less than 76.4% nucleotide identity across the spike protein.
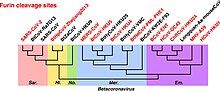
[28] SARS-CoV-2 shares amino acid identity with a furin cleavage site of human ENaC α subunit.
[35] Within a single host, a variety of single-nucleotide variations arise through random mutations and genetic drift giving rise to viral quasispecies.
[35][37] The process of host adaptation has been studied in humanized mice as well as by generating mouse-adapted viral strains through serial passage.
[47] Tai et al. wrote that population expansion rather than positive selection explained the mutation frequency spectrum during the early pandemic.
[49] Cagliani et al. wrote that the SARS-CoV-2 genome overall shows evidence of "strong to moderate" purifying selection.
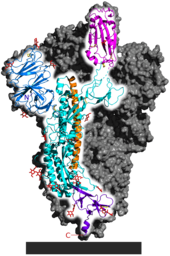
[51] Strong evidence of positive selection was found however in the spike protein S1 subunit, which contains the receptor binding domain.
[43] The receptor binding domain is a significant factor in host tropism, or the variety of species a virus can infect.
[55] Kang et al. identified a single nucleotide polymorphism relative to RaTG13 in the spike protein, consistent among all of more than 180,000 SARS-CoV-2 samples, affecting glycosylation of the receptor binding domain.
[56] Using a reverse genetics system to generate an ancestral-like mutant, they confirmed that the putative ancestral form of this SNP was much less transmissible in human cells.
[59] Guo et al. identified clade 1 and 2 sarbecoviruses in Rhinolophus fecal samples, suggesting that both types naturally replicate in the bat digestive tract.
[63] Graham and Baric wrote that in the case of SARS-CoV-1, virus populations circulated and adapted to civets and humans over the course of two years before the recognized outbreak.
[54] In the outbreak of SARS-CoV-1, palm civets, raccoon dogs, ferret badgers, red foxes, domestic cats, and rice field rats were possible vectors.
[7] Graham and Baric wrote that human and civet infections likely stemmed from an unknown common progenitor.
[69] Patrick Berche wrote that the emergences of SARS-CoV-1 and MERS-CoV appeared to be sequential processes involving intermediate hosts, co-infections, and recombination.
[19] Frutos et al. proposed that rather than a discrete spillover event, SARS-CoV-2 arose in accordance with a circulation model, involving repeated horizontal transfer among humans, bats, and other mammals without establishing significant reservoirs in any of them until the pandemic.
[73] Pangolins are sometimes sold in wet markets in China, where they are considered a culinary delicacy and a component of traditional medicine.
[13] The highest sequence similarity to the SARS-CoV-2 spike receptor binding domain was found in a coronavirus infecting Sunda pangolins in Guangdong province.
[77] Wild and semi-wild game animals are commonly traded and consumed in China, and this practice has expanded in recent decades.
[80] In congressional testimony, coronavirus expert Ralph Baric stated that "the market was the site of amplification in late December, January.
[85] Overall, wildlife including racoon dogs were detected at very low levels and mostly associated with negative samples.
[86] In particular, they wrote that the evidence does not prove the presence of an infected raccoon dog or the occurrence of multiple zoonotic spillovers at the market as proposed by Pekar et al.
[55] Theories for the origin of the omicron variant include long-term circulation among humans outside areas where genetic surveillance was performed, mutation in an immunocompromised individual, or adaptation in an animal species after reverse zoonosis.