Cancer epigenetics
[6][7] In different types of cancer, a variety of epigenetic mechanisms can be perturbed, such as the silencing of tumor suppressor genes and activation of oncogenes by altered CpG island methylation patterns, histone modifications, and dysregulation of DNA binding proteins.
[13] Hypomethylation of CpG dinucleotides in other parts of the genome leads to chromosome instability due to mechanisms such as loss of imprinting and reactivation of transposable elements.
[22] Additionally, mouse models have shown that a decrease in histone H4R3 asymmetric dimethylation (H4R3me2a) of the p19ARF promoter is correlated with more advanced cases of tumorigenesis and metastasis.
[25][26] Some research has focused on blocking the action of BRD4 on acetylated histones, which has been shown to increase the expression of the Myc protein, implicated in several cancers.
[28] CTCF, or CCCTC binding factor, is a zinc finger protein that insulates the p53 promoter from accumulating repressive histone marks.
In certain types of cancer cells, the CTCF protein does not bind normally, and the p53 promoter accumulates repressive histone marks, causing p53 expression to decrease.
The histone variants of the H2A family are highly conserved in mammals, playing critical roles in regulating many nuclear processes by altering chromatin structure.
A new emerging field that captures toxicological epigenetic changes as a result of the exposure to different compounds (drugs, food, and environment) is toxicoepigenetics.
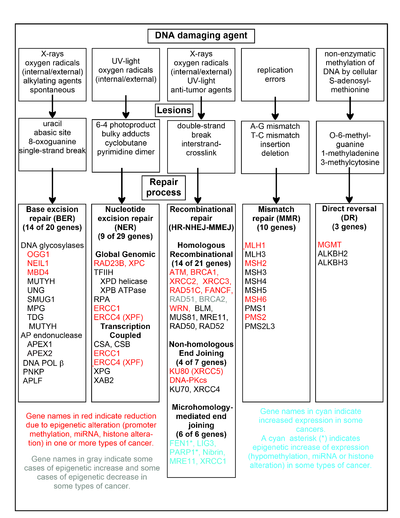
[31] DNA damage, caused by UV light, ionizing radiation, environmental toxins, and metabolic chemicals, can also lead to genomic instability and cancer.
[37] Dysregulation of metabolism allows tumor cells to generate needed building blocks as well as to modulate epigenetic marks to support cancer initiation and progression.
Cancer-induced metabolic changes alter the epigenetic landscape, especially modifications on histones and DNA, thereby promoting malignant transformation, adaptation to inadequate nutrition, and metastasis.
In order to satisfy the biosynthetic demands of cancer cells, metabolic pathways are altered by manipulating oncogenes and tumor suppressive genes concurrently.
More recently, Tessitore et al.[45] listed further DNA repair genes that are directly targeted by additional miRNAs, including ATM (miR-18a, miR-101), DNA-PK (miR-101), ATR (miR-185), Wip1 (miR-16), MLH1, MSH2 and MSH6 (miR-155), ERCC3 and ERCC4 (miR-192) and UNG2 (mir-16, miR-34c and miR-199a).
[47] However, the majority of 68 sporadic colon cancers with reduced expression of the DNA mismatch repair protein MLH1 were found to be deficient due to epigenetic methylation of the CpG island of the MLH1 gene.
Human neoplasias, including thyroid, prostatic, cervical, colorectal, pancreatic and ovarian carcinoma, show a strong increase of HMGA1a and HMGA1b proteins.
[62] FEN1, the flap endonuclease in MMEJ, is epigenetically increased by promoter hypomethylation and is over-expressed in the majority of cancers of the breast,[63] prostate,[64] stomach,[65][66] neuroblastomas,[67] pancreas,[68] and lung.
Epigenetic defects in DNA repair may thus contribute to the characteristic high frequency of mutations in the genomes of cancers, and cause their carcinogenic progression.
In an early study, looking at a limited set of transcriptional promoters, Fernandez et al.[94] examined the DNA methylation profiles of 855 primary tumors.
Additionally, increased CpG site methylation was found in low levels in most of the five host nuclear genes studied, including 5/5 TERT, 1/4 DAPK1, 2/5 RARB, MAL, and CADM1.
[109] Mutations in MLL block the correct regulatory regions in leukemia associated translocations or insertions causing malignant transformation controlled by HOX genes.
p16 is a tumor suppressor protein that occurs in mostly in humans the functional significance of the mutations was tested on many other species including mice, cats, dogs, monkeys and cows the identification of these multiple nonoverlapping clones was not entirely surprising since the reduced stringency hybridization of a zoo blot with the same probe also revealed 10-15 positive EcoRI fragments in all species tested.
In primary tumor and mediastinal lymph node biopsy samples, hypermethylation of both CDKN2A and CDH13 serves as the marker for increased risk of faster cancer relapse and higher death rate of patients.
[126] Epigenetic control of the proto-onco regions and the tumor suppressor sequences by conformational changes in histones plays a role in the formation and progression of cancer.
[130] Drugs that specifically target the inverted methylation pattern of cancerous cells include the DNA methyltransferase inhibitors azacitidine[131][132] and decitabine.
[133][134] These hypomethylating agents are used to treat myelodysplastic syndrome,[135] a blood cancer produced by abnormal bone marrow stem cells.
[12] These agents inhibit all three types of active DNA methyltransferases, and had been thought to be highly toxic, but proved to be effective when used in low dosage, reducing progression of myelodysplastic syndrome to leukemia.
The ability for epigenetic mechanisms to be reversed is attributed to the fact that the coding of the genes being silenced through histone and DNA modification is not being altered.
[145] The goal for epigenetic therapies is to repress this methylation and reverse these modifications in order to create a new epigenome where cancer cells no longer thrive and tumor suppression is the new function.
Synthetic drugs are used as tools in epigenetic therapies due to their ability to inhibit enzymes causing histone modifications and DNA methylations.
The Food and Drug Administration (FDA) has currently approved one hypomethylating agent which, through the conduction of clinical trials, has shown promising results when utilized to treat patients with Myelodysplastic Syndrome (MDS).