Chloroplast DNA
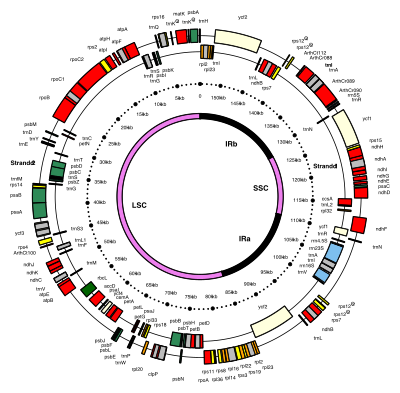
[9] The inverted repeats vary wildly in length, ranging from 4,000 to 25,000 base pairs long each.
[11] Inverted repeats in plants tend to be at the upper end of this range, each being 20,000–25,000 base pairs long.
[11] While a given pair of inverted repeats are rarely completely identical, they are always very similar to each other, apparently resulting from concerted evolution.
[11] The inverted repeat regions are highly conserved among land plants, and accumulate few mutations.
[18] The first chloroplast genomes were sequenced in 1986, from tobacco (Nicotiana tabacum)[19] and liverwort (Marchantia polymorpha).
[21][22] It also demonstrated the significant extent of gene transfer from the cyanobacterial ancestor to the nuclear genome.
[23][24] The genes primarily encode core components of the photosynthetic machinery and factors involved in their expression and assembly.
[25] Across species of land plants, the set of genes encoded by the chloroplast genome is fairly conserved.
[25] The large Rubisco subunit and 28 photosynthetic thylakoid proteins are encoded within the chloroplast genome.
[32] In land plants, some 11–14% of the DNA in their nuclei can be traced back to the chloroplast,[33] up to 18% in Arabidopsis, corresponding to about 4,500 protein-coding genes.
[34] There have been a few recent transfers of genes from the chloroplast DNA to the nuclear genome in land plants.
[37] RNA editing is the insertion, deletion, and substitution of nucleotides in a mRNA transcript prior to translation to protein.
The highly oxidative environment inside chloroplasts increases the rate of mutation so post-transcription repairs are needed to conserve functional sequences.
These proteins consist of 35-mer repeated amino acids, the sequence of which determines the cis binding site for the edited transcript.
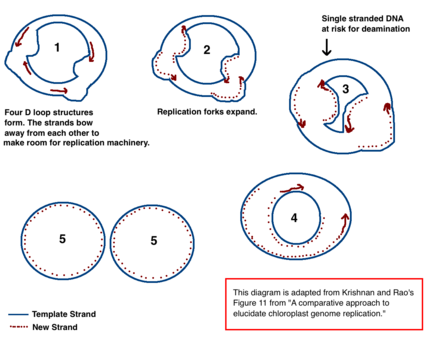
[39] The mechanism for chloroplast DNA (cpDNA) replication has not been conclusively determined, but two main models have been proposed.
[40][41] The results of the microscopy experiments led to the idea that chloroplast DNA replicates using a double displacement loop (D-loop).
It further contends that only a minority of the genetic material is kept in circular chromosomes while the rest is in branched, linear, or other complex structures.
[43] Curiously, around half of the protein products of transferred genes aren't even targeted back to the chloroplast.
Many became exaptations, taking on new functions like participating in cell division, protein routing, and even disease resistance.
[48] N-terminal transit sequences are also called presequences[43] because they are located at the "front" end of a polypeptide—ribosomes synthesize polypeptides from the N-terminus to the C-terminus.
[45] Chloroplast transit peptides exhibit huge variation in length and amino acid sequence.
[43] After a chloroplast polypeptide is synthesized on a ribosome in the cytosol, ATP energy can be used to phosphorylate, or add a phosphate group to many (but not all) of them in their transit sequences.
[43] Serine and threonine (both very common in chloroplast transit sequences—making up 20–30% of the sequence)[50] are often the amino acids that accept the phosphate group.
[48][50] The enzyme that carries out the phosphorylation is specific for chloroplast polypeptides, and ignores ones meant for mitochondria or peroxisomes.
[46] Toc34 is an integral protein in the outer chloroplast membrane that's anchored into it by its hydrophobic[53] C-terminal tail.
[51] Toc34's job is to catch some chloroplast preproteins in the cytosol and hand them off to the rest of the TOC complex.
[43] When GTP, an energy molecule similar to ATP attaches to Toc34, the protein becomes much more able to bind to many chloroplast preproteins in the cytosol.
[43] The chloroplast preprotein's presence causes Toc34 to break GTP into guanosine diphosphate (GDP) and inorganic phosphate.
This suggests that it might act as a shuttle that finds chloroplast preproteins in the cytosol and carries them back to the TOC complex.
Tic100 is found at the edges of the 1 million dalton complex on the side that faces the chloroplast intermembrane space.