Circular chromosome
The E. coli origin of replication, called oriC consists of DNA sequences that are recognised by the DnaA protein, which is highly conserved amongst different bacterial species.
DnaA binding to the origin initiates the regulated recruitment of other enzymes and proteins that will eventually lead to the establishment of two complete replisomes for bidirectional replication.
The methylation of adenines is important as it alters the conformation of DNA to promote strand separation,[6] and it appears that this region of oriC has a natural tendency to unwind.
[8] After DnaB translocates to the apex of each replication fork, the helicase both unwinds the parental DNA and interacts momentarily with primase.
In addition, DNA gyrase is needed to relieve the topological stress created by the action of DnaB helicase.
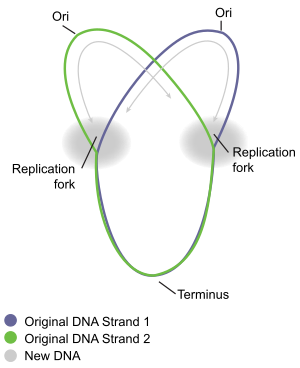
When the replication fork moves around the circle, a structure shaped like the Greek letter theta Ө is formed.
He then isolated the chromosomes by lysing the cells gently and placed them on an electron micrograph (EM) grid which he exposed to X-ray film for two months.
The regions of DNA undergoing replication during the experiment were then visualized by using autoradiography and examining the developed film microscopically.
[12] The E. coli DNA polymerase III holoenzyme is a 900 kD complex, possessing an essentially a dimeric structure.
Deoxynucleotides are then added to this primer by a single DNA polymerase III dimer, in an integrated complex with DnaB helicase.
Leading strand synthesis then proceeds continuously, while the DNA is concurrently unwound at the replication fork.
[15] The encountered DNA damages are ordinarily processed by recombinational repair enzymes to allow continued replication fork progression.
[15] Termination is the process of fusion of replication forks and disassembly of the replisomes to yield two separate and complete DNA molecules.
The arrangement of the Ter sites forms two opposed groups that forces the two forks to meet each other within the region they span.
In an experiment by Zechiedrich, Khodursky and Cozzarelli in 1997, it was found that topoisomerase IV is the only important decatenase of DNA replication intermediates in bacteria.
This suggests that Topoisomerase IV is the primary protein for decatenation of interlinked chromosomes in vivo, with DNA gyrase playing a minor role.