Conserved signature indels
Conserved signature inserts and deletions (CSIs) in protein sequences provide an important category of molecular markers for understanding phylogenetic relationships.
[1][2] CSIs, brought about by rare genetic changes, provide useful phylogenetic markers that are generally of defined size and they are flanked on both sides by conserved regions to ensure their reliability.
[2][3][4][5] The CSIs that are restricted to a particular clade or group of species, generally provide good phylogenetic markers of common evolutionary descent.
[2] Due to the rarity and highly specific nature of such changes, it is less likely that they could arise independently by either convergent or parallel evolution (i.e. homoplasy) and therefore are likely to represent synapomorphy.
[2][3] By determining the presence or absence of CSIs in an out-group species, one can infer whether the ancestral form of the CSI was an insert or deletion and this can be used to develop a rooted phylogenetic relationship among organisms.
[3] Compared to tree branching orders which can vary among methods, specific CSIs make for more concrete circumscriptions that are computationally cheaper to apply.
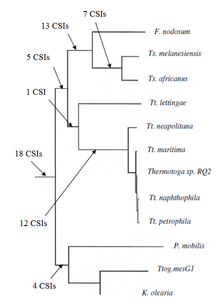
[6] Group-specific CSIs are commonly shared by different species belonging to a particular taxon (e.g. genus, family, class, order, phylum) but they are not present in other groups.
[7] Group-specific CSIs have been used in the past to determine the phylogenetic relationship of a number of bacterial phyla and subgroups within it.
This signature indicates a specific relationship of taxa X, Y and Z and also A, B and C. Based upon the presence or absence of such an indel, in out-group species (viz.
[7] Mainline CSIs have been used in the past to determine the phylogenetic relationship of a number of bacterial phyla.
[12][13][14][15][16] However in recent years the discovery and analyses of conserved indels (CSIs) in many universally distributed proteins have aided in this quest.
A detailed phylogenetic study using the CSI approach was conducted to distinguish these phyla in molecular terms.
6 CSIs were uniquely found in various Nitrososphaerota, namely Cenarchaeum symbiosum, Nitrosopumilus maritimus and a number of uncultured marine Thermoproteota.
The signatures described provide novel means for distinguishing Thermoproteota and Nitrososphaerota, additionally they could be used as a tool for the classification and identification of related species.
Firstly, a phylogenetic tree based on concatenated sequences of a number of universally-distributed proteins was created.