Coronavirus nucleocapsid protein
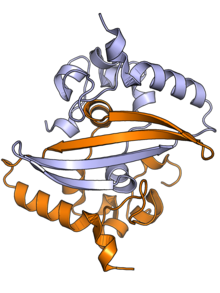
[2][3] A third small domain at the C-terminal tail appears to have an ordered alpha helical secondary structure and may be involved in the formation of higher-order oligomeric assemblies.
[2] Parts of the IDR, particularly a conserved sequence motif rich in serine and arginine residues (the SR-rich region), may also be implicated in dimer formation, though reports on this vary.
[2][3] Although higher-order oligomers formed through the C-terminal domain have been observed crystallographically, it is unclear if these structures have a physiological role.
Type I PRMT inhibitor (MS023) or substitution of R95 or R177 with lysine inhibited interaction of N protein with the 5’-UTR of SARS-CoV-2 genomic RNA, a property required for viral packaging | doi: 10.1016/j.jbc.2021.100821 | PMID 34029587.
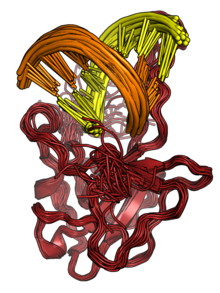
[4] The N protein binds to RNA to form ribonucleoprotein (RNP) structures for packaging the genome into the viral capsid.
[2][3] The RNP particles formed are roughly spherical and are organized in flexible helical structures inside the virus.
[2][3] Formation of RNPs is thought to involve allosteric interactions between RNA and multiple RNA-binding regions of the protein.
N is physically colocalized with the viral RNA-dependent RNA polymerase early in the replication cycle and forms interactions with non-structural protein 3, a component of the replicase-transcriptase complex.
In several coronaviruses, including SARS-CoV, the N protein has been reported to cause cell cycle arrest in S phase through interactions with cyclin-CDK.
[24] The N protein's properties of being well conserved, not appearing to recombine frequently, and producing a strong T-cell response have led to it being studied as a potential target for coronavirus vaccines.