Developmental bias
[1][2] Historically, the term was synonymous with developmental constraint,[1][3][4] however, the latter has been more recently interpreted as referring solely to the negative role of development in evolution.
[6][page needed] In the Structuralist view, phenotypic evolution is the result of the action of natural selection on previously ‘filtered’ variation during the course of ontogeny.
[7][8] It contrasts with the Functionalist (also “adaptationist”, “pan-selectionist” or “externalist”) view in which phenotypic evolution results only from the interaction between the deterministic action of natural selection and variation caused by mutation.
[3][7] The rationale behind the role of the organism, or more specifically the embryo, as a causal force in evolution and for the existence of bias is as follows: The traditional, neo-Darwinian, approach to explain the process behind evolutionary change is natural selection acting upon heritable variation caused by genetic mutations.
[6] For a mutation to readily alter a phenotype, and hence be visible to natural selection, it has to modify the ontogenetic trajectory, a process referred to as developmental reprogramming.
[10] Some kinds of reprogramming are more likely to occur than others given the nature of the genotype–phenotype map, which determines the propensity of a system to vary in a particular direction,[8][11] thus, creating a bias.
[14][5][6] This type of bias is thought to facilitate adaptive evolution by aligning phenotypic variability with the direction of selection.
[8] An important distinction between structuralism and functionalism regards primarily with the interpretation of the causes of the empty regions in the morphospace (that is, the inexistent phenotypes): Under the functionalist view, empty spaces correspond to phenotypes that are both ontogenetically possible and equally probable but are eliminated by natural selection due to their low fitness.
[7][24] This trend was referred to as transpecific parallelism, suggesting the existence of profound historical rules governing the expression of abnormal forms in distantly related species.
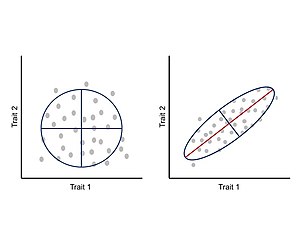
[30][31] Thus, a population’s immediate ability to respond to selection is determined by the G-matrix, in which the variance is a function of standing genetic variation, and the covariance arises from pleiotropy and linkage disequilibrium.
In other words, if the main axis of variation is aligned with the direction of selection, covariation (genetic or phenotypic) will facilitate the rate of adaptive evolution; however, if the main axis of variation is orthogonal to the direction of selection, covariation will constraint the rate of adaptive evolution.
[2][12][25] In general, for a population under the influence of a single fitness optimum, the rate of morphological divergence (from an ancestral to a new phenotype or between pairs of species) is inversely proportional to the angle formed by the main axis of variation and the direction of selection, causing a curved trajectory through the morphospace.
[19][36] In general, it is expected that newly arisen mutations with higher dominance and fewer pleiotropic and epistatic effects are more likely to be targets of evolution,[37] thus, the hierarchical architecture of developmental pathways may bias the genetic basis of evolutionary change.
[6][41] In development, neutral networks are clusters of GRNs that differ in only one interaction between two nodes (e.g. replacing transcription with suppression) and yet produce the same phenotypic outcome.