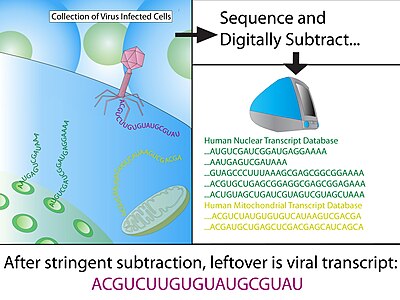
Digital transcriptome subtraction
Digital transcriptome subtraction (DTS) is a bioinformatics method to detect the presence of novel pathogen transcripts through computational removal of the host sequences.
DTS is the direct in silico analogue of the wet-lab approach representational difference analysis (RDA), and is made possible by unbiased high-throughput sequencing and the availability of a high-quality, annotated reference genome of the host.
In a proof of principle experiment, Meyerson et al. demonstrated that it was a feasible approach using Epstein–Barr virus-infected lymphocytes in post-transplant lymphoproliferative disorder (PTLD).
[3] In 2007, the term "Digital Transcriptome Subtraction" was coined by the Chang-Moore group,[4] and was used to discover Merkel cell polymavirus in Merkel-cell carcinoma.
[1] Simultaneously to the MCV discovery, this approach was used to implicate a novel arenavirus as cause of fatality in a case where three patients died of similar illnesses shortly following organ transplantations from a single donor.
Messenger RNA is then purified using an oligo-dT column that binds to the poly-A tail, a signal specifically found on transcribed genes.