Drosophila circadian rhythm
[2] Drosophila circadian rhythm was discovered in 1935 by German zoologists, Hans Kalmus and Erwin Bünning.
American biologist Colin S. Pittendrigh provided an important experiment in 1954, which established that circadian rhythm is driven by a biological clock.
The genetics was first understood in 1971, when Seymour Benzer and Ronald J. Konopka reported that mutation in specific genes changes or stops the circadian behaviour.
[3] During the process of eclosion by which an adult fly emerges from the pupa, Drosophila exhibits regular locomotor activity (by vibration) that occurs during 8-10 hours intervals starting just before dawn.
[5] After almost two decades, the existence of the circadian clock was discovered by Seymour Benzer and his student Ronald J. Konopka at the California Institute of Technology.
The main flight activity generally takes place in the morning and another peak occurs before sunset.
In 1980, Jeffrey C. Hall and his student Charalambos P. Kyriacou, at Brandeis University in Waltham, discovered that courtship activity is also controlled by per gene.
[12][13] At the same time, Michael W. Young's team at the Rockefeller University reported similar effects of per, and that the gene covers 7.1-kilobase (kb) interval on the X chromosome and encodes a 4.5-kb poly(A)+ RNA.
[14][15] In 1986, they sequenced the entire DNA fragment and found the gene encodes the 4.5-kb RNA, which produces a protein, a proteoglycan, composed of 1,127 amino acids.
[21] In the late 1998, Hall and Roshbash's team discovered cryb, a gene for sensitivity to blue light.
The activity of cry is under circadian regulation, and influenced by other genes such as per, tim, clk, and cyc.
[23] The gene product CRY is a major photoreceptor protein belonging to a class of flavoproteins called cryptochromes.
[24] In 1998, Hall and Jae H. Park isolated a gene encoding a neuropeptide named pigment dispersing factor (PDF), based on one of the roles it plays in crustaceans.
[26] In 2001, Young and his team demonstrated that glycogen synthase kinase-3 (GSK-3) ortholog shaggy (SGG) is an enzyme that regulates TIM maturation and accumulation in the early night, by causing phosphorylation.
[27] Hall, Rosbash, and Young shared the Nobel Prize in Physiology or Medicine 2017 “for their discoveries of molecular mechanisms controlling the circadian rhythm”.
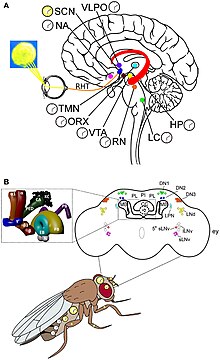
[33] Drosophila circadian keeps time via daily fluctuations of clock-related proteins which interact in a transcription-translation feedback loop.
PER and TIM proteins are synthesized in the cytoplasm and exhibit a smooth increase in levels over the day.
The actual degradation is through the ubiquitin-proteasome pathway and is carried out by a ubiquitin ligase called Slimb (supernumerary limbs).
[37] In the early morning, the appearance of light causes PER and TIM proteins to break down in a network of transcriptional activation and repression.