Electron-transfer dissociation
[3] Transferring an electron causes peptide backbone cleavage into c- and z-ions while leaving labile post translational modifications (PTM) intact.
[7] Because ECD requires a large amount of near-thermal electrons (<0.2eV), originally it was used exclusively with Fourier transform ion cyclotron resonance mass spectrometry (FTICR), the most expensive form of MS instrumentation.
[9] In 2004 Syka et al. announced the creation of ETD, a dissociation method similar to ECD, but using a low-cost, widely available commercial spectrometer.
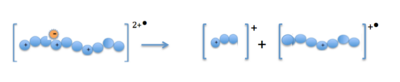
In order for an electron to be transferred to the positive precursor molecules radical anions are generated and put into the ion trap with them.
[15] Sequences of 15-40 amino acids at both the N-terminus and the C-terminus of the protein can be read using mass-to-charge values for the singly and doubly charged ions.
[16] Electron transfer dissociation takes place in an ion trap mass spectrometer with an electrospray ionization source.
The first ETD experiments at the University of Virginia utilized a radio frequency quadrupole linear ion trap (LQT) modified with a chemical ionization (CI) source at the back side of the instrument (see diagram at right).
Having a negative CI source at the back of the instrument interfered with the high-resolution analyzer in LQT-Orbitrap and quadrupole time-of-flight (QTOF), so alternate ionization methods for the radical anions have been introduced.
[17] Later a lab at the University of Wisconsin adapted a hybrid quadrupole linear ion trap-orbitrap mass spectrometer to use ETD.