Fixation index
The fixation index (FST) is a measure of population differentiation due to genetic structure.
Developed as a special case of Wright's F-statistics, it is one of the most commonly used statistics in population genetics.
A zero value implies complete panmixia; that is, that the two populations are interbreeding freely.
Also, strictly speaking FST is not a distance in the mathematical sense, as it does not satisfy the triangle inequality.
Two of the most commonly used definitions for FST at a given locus are based on 1) the variance of allele frequencies among populations, and on 2) the probability of identity by descent.
is the variance of the allelic state in the total population, FST is defined as [2] Wright's definition illustrates that FST measures the amount of genetic variance that can be explained by population structure.
Using this definition, FST can be interpreted as measuring how much closer two individuals from the same subpopulation are, compared to the total population.
If the mutation rate is small, this interpretation can be made more explicit by linking the probability of identity by descent to coalescent times: Let T0 and T denote the average time to coalescence for individuals from the same subpopulation and the total population, respectively.
However, this estimator is biased when sample sizes are small or if they vary between populations.
Two of the most widely used procedures are the estimator by Weir & Cockerham (1984),[5] or performing an Analysis of molecular variance.
Closely related ethnic groups, such as the Danes vs. the Dutch, or the Portuguese vs. the Spaniards show values significantly below 1%, indistinguishable from panmixia.
Within Europe, the most divergent ethnic groups have been found to have values of the order of 7% (Sámi vs. Sardinians).
[6] A genetic distance of 0.125 implies that kinship between unrelated individuals of the same ancestry relative to the world population is equivalent to kinship between half siblings in a randomly mating population.
Their initial database contains 76,676 gene frequencies (using 120 blood polymorphisms), corresponding to 6,633 samples in different locations.
They focus on aboriginal populations that were at their present location at the end of the 15th century when the great European migrations began.
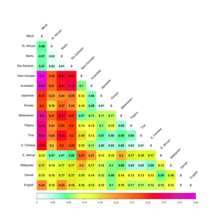
For these 42 populations, Cavalli-Sforza and coauthors report bilateral distances computed from 120 alleles.
When considering more disaggregated data for 26 European populations, the smallest genetic distance (0.0009) is between the Dutch and the Danes, and the largest (0.0667) is between the Lapps and the Sardinians.
The mean genetic distance among the 861 available pairings of the 42 selected populations was found to be 0.1338.[page needed].
The following table shows Fst calculated by Cavalli-Sforza (1994) for some populations: A 2012 study based on International HapMap Project data estimated FST between the three major "continental" populations of Europeans (combined from Utah residents of Northern and Western European ancestry from the CEPH collection and Italians from Tuscany), East Asians (combining Han Chinese from Beijing, Chinese from metropolitan Denver and Japanese from Tokyo, Japan) and Sub-Saharan Africans (combining Luhya of Webuye, Kenya, Maasai of Kinyawa, Kenya and Yoruba of Ibadan, Nigeria).
[9] Pairwise Fst values among several populations based on whole exome sequencing (WES) in 2016:[12]