Glutamine synthetase
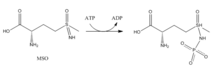
[4] The amide group of glutamate is a nitrogen source for the synthesis of glutamine pathway metabolites.
[6][7] Ammonium ion binds more strongly than water to GS due to electrostatic forces between a cation and a negatively charged pocket.
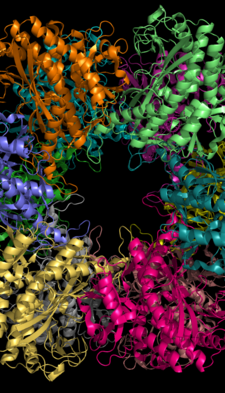
The C-terminus (helical thong) stabilizes the GS structure by inserting into the hydrophobic region of the subunit across in the other ring.
In addition, the central channel is formed via six four-stranded β-sheets composed of anti-parallel loops from the twelve subunits.
[5][7] The presence of ADP causes a conformational shift in GS that stabilizes the γ-glutamyl phosphate moiety.
[7] In the second step, deprotonation of ammonium allows ammonia to attack the intermediate from its nearby site to form glutamine.
One study shows that morphological changes occur that increase GS expression in glutamatergic areas or other adaptations that alleviates high levels of glutamate and ammonia.
[25] Adenylylation is a post-translational modification involving the covalent attachment of AMP to a protein side chain.
[25] The AT:PIIA and AT:PIID complexes are allosterically regulated in a reciprocal fashion by α-ketoglutarate (α-KG) and glutamine (Gln).
[25] In the majority of gram-negative bacteria, GS can be modified by adenylylation (some cyanobacteria and green algae or exceptions).
[6] Other inhibitors are the result of glutamine metabolism: tryptophan, histidine, carbamoyl phosphate, glucosamine-6-phosphate, cytidine triphosphate (CTP), and adenosine monophosphate (AMP).
[6] L-serine, L-alanine, and glycine bind to the site for L-glutamate in unadenylated GS.
[8] Many inhibitory input signals allows for fine tuning of GS by reflecting nitrogen levels in the organism.
Feedback regulation distinguishes the difference between two eukaryotic types of GS: brain and non-brain tissues.
If NRII is complexed with PIIA then it will function as a phosphatase and NRI-P is converted back to NRI.
[28] Instead of the common NtrC-NtrB two component system,[29][30] cyanobacteria harbour the transcriptional regulator NtcA which is restricted to this clade and controls expression of GS and a multitude of genes involved in nitrogen metabolism.
[35] In addition, expression of the GS inactivating factor IF17 is controlled by a glutamine-binding riboswitch.