Hammerhead ribozyme
Although the cleavage takes place in the absence of protein enzymes, the hammerhead RNA itself is not a catalyst in its natural state, as it is consumed by the reaction (i.e. performs self-cleavage) and therefore cannot catalyze multiple turnovers.
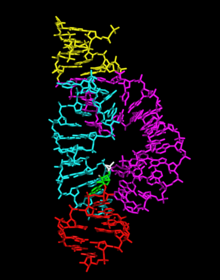
The minimal trans-acting hammerhead ribozyme sequence that is catalytically active consists of three base-paired stems flanking a central core of 15 conserved (mostly invariant) nucleotides, as shown.
Its small size, thoroughly-investigated cleavage chemistry, known crystal structure, and its biological relevance make the hammerhead ribozyme particularly well-suited for biochemical and biophysical investigations into the fundamental nature of RNA catalysis.
Hammerhead ribozymes may play an important role as therapeutic agents; as enzymes which tailor defined RNA sequences, as biosensors, and for applications in functional genomics and gene discovery.
[10] New examples of this ribozyme were then found in the genomes of unrelated organisms like schistosomes,[11] cave crickets,[12] Arabidopsis thaliana [13] and a few mammals like rodents and the platypus.
[16] Similar reports confirmed and extended these observations,[17][18][19] unveiling the hammerhead ribozyme as a ubiquitous catalytic RNA in all life kingdoms.
[20] Most eukaryotic hammerhead ribozymes are related to a kind of short interspersed retroelements (SINEs) called retrozymes, which express as small circular RNAs.
[24] In 2021, novel Hepatitis D virus genomes of circular RNA from diverse animals were found to encode hammerhead ribozymes similar to those present in plant viroids and viral satellites.
[26] The hammerhead ribozyme carries out a very simple chemical reaction that results in the breakage of the substrate strand of RNA, specifically at C17, the cleavage-site nucleotide.
The attacking and leaving group oxygens will both occupy the two axial positions in the trigonal bipyramidal transition-state structure as is required for an SN2-like reaction mechanism.
The reaction is therefore reversible, as the scissile phosphate remains a phosphodiester, and may thus act as a substrate for hammerhead RNA-mediated ligation without a requirement for ATP or a similar exogenous energy source.
It was assumed that divalent metal ions like Mg2+ were thought to have two roles: Promote proper folding of RNA and to form the catalytic core.
This discovery suggested that the RNA itself, rather than serving as an inert, passive scaffold for the binding of chemically active divalent metal ions, is instead itself intimately involved in the chemistry of catalysis.
The minimal hammerhead sequence that is required for the self-cleavage reaction includes approximately 13 conserved or invariant "core" nucleotides, most of which are not involved in forming canonical Watson-Crick base-pairs.
The core region is flanked by Stems I, II and III, which are in general made of canonical Watson-Crick base-pairs but are otherwise not constrained with respect to sequence.
The catalytic turnover rate of minimal hammerhead ribozymes is ~ 1/min (a range of 0.1/min to 10/min is commonly observed, depending upon the nonconserved sequences and the lengths of the three helical stems) under standard reaction conditions of high Mg2+ (~10 mM), pH 7.5 and 25 °C.
[31][32] The minimal hammerhead ribozyme has been exhaustively studied by biochemists and enzymologists as well as by X-ray crystallographers, NMR spectroscopists, and other practitioners of biophysical techniques.
[34] The minimal hammerhead ribozyme is composed of three base paired helices, separated by short linkers of conserved sequence as shown in the crystal structure.
For example, the invariant core residues G5, G8, G12 and C3 in the minimal hammerhead ribozyme were each observed to be so fragile that changing even a single exocyclic functional group on any one of these nucleotides results in a dramatic reduction or abolition of catalytic activity, yet few of these appeared to form hydrogen bonds involving the Watson-Crick faces of these nucleotide bases in any of the minimal hammerhead structures, apart from a G-5 interaction in the product structure.
Perhaps most worrisome were experiments that suggested the A-9 and scissile phosphates must come within about 4 Å of one another in the transition-state, based upon double phosphorothioate substitution and soft metal ion rescue experiments; the distance between these phosphates in the minimal hammerhead crystal structure was about 18 Å, with no clear mechanism for close approach if the Stem II and Stem I A-form helices were treated as rigid bodies.
Taken together, these results appeared to suggest that a fairly large-scale conformational change must have taken place in order to reach the transition-state within the minimal hammerhead ribozyme structure.
For these reasons, the two sets of experiments (biochemical vs. crystallographic) appeared not only to be at odds, but to be completely and hopelessly irreconcilable, generating a substantial amount of discord in the field.
This is probably the source of the observation that divalent metal ions are required at low ionic strength, but can be dispensed with at higher concentrations of monovalent cations.
hammerhead ribozyme RNA molecule





