Hox gene
For example, Hox genes in insects specify which appendages form on a segment (for example, legs, antennae, and wings in fruit flies), and Hox genes in vertebrates specify the types and shape of vertebrae that will form.
Studies on Hox genes in ciliated larvae have shown they are only expressed in future adult tissues.
In larvae with complete metamorphosis the Hox genes are mainly expressed in juvenile rudiments and are absent in the transient larval tissues.
[1][2] An analogy for the Hox genes can be made to the role of a play director who calls which scene the actors should carry out next.
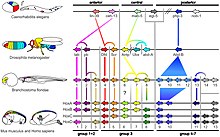
[10][13] In most bilaterian animals, Hox genes are expressed in staggered domains along the head-to-tail axis of the embryo, suggesting that their role in specifying position is a shared, ancient feature.
The general principles of Hox gene function and logic elucidated in flies will apply to all bilaterian organisms, including humans.
A dominant Antp mutation, caused by a chromosomal inversion, causes Antp to be expressed in the antennal imaginal disc, so that, instead of forming an antenna, the disc makes a leg, resulting in a leg coming out of the fly's head.
One of the many genes that Ubx represses is blistered, which activates proteins involved in cell-cell adhesion, and spalt, which patterns the placement of wing veins.
Regulatory abd-B suppress embryonic ventral epidermal structures in the eighth and ninth segments of the Drosophila abdomen.
[18] Proteins with a high degree of sequence similarity are also generally assumed to exhibit a high degree of functional similarity, i.e. Hox proteins with identical homeodomains are assumed to have identical DNA-binding properties (unless additional sequences are known to influence DNA-binding).
For Hox proteins, three different classification schemes exist: phylogenetic inference based, synteny-based, and sequence similarity-based.
[22] The three classification schemes provide conflicting information for Hox proteins expressed in the middle of the body axis (Hox6-8 and Antp, Ubx and abd-A).
The base pairs following this initial sequence are used to distinguish between homeodomain proteins, all of which have similar recognition sites.
For instance, the nucleotide following the TAAT sequence is recognized by the amino acid at position 9 of the homeodomain protein.
In the maternal protein Bicoid, this position is occupied by lysine, which recognizes and binds to the nucleotide guanine.
If the lysine in Bicoid is replaced by glutamine, the resulting protein will recognize Antennapedia-binding enhancer sites.
One of these, HOTAIR, silences in trans (it is transcribed from the HOXC cluster and inhibits late HOXD genes) by binding to Polycomb-group proteins (PRC2).
[35] In higher animals including humans, retinoic acid regulates differential expression of Hox genes along the anteroposterior axis.
[36] Genes in the 3' ends of Hox clusters are induced by retinoic acid resulting in expression domains that extend more anteriorly in the body compared to 5' Hox genes that are not induced by retinoic acid resulting in expression domains that remain more posterior.
In some organisms, especially vertebrates, the various Hox genes are situated very close to one another on the chromosome in groups or clusters.
The Hox genes are named for the homeotic phenotypes that result when their function is disrupted, wherein one segment develops with the identity of another (e.g. legs where antennae should be).
HOX genes control the regulation and development of many key structures in the body, such as somites, which form the vertebrae and ribs, the dermis of the dorsal skin, the skeletal muscles of the back, and the skeletal muscles of the body wall and limbs.
HOX genes help differentiate somite cells into more specific identities and direct them to develop differently depending on where they are in the body.
Due to the fact that the HOX genes are so highly conserved, most research has been done on much simpler model organisms, such as mice.
One of the major differences that was noticed when comparing mice and drosophila, in particular, has to do with the location and layering of HOX genes within the genome.
[50] This rapid evolvability is in part because invertebrates experienced much more dramatic episodes of adaptive radiation and mutations.
After the rediscovery of Mendel's genetic principles, Bateson and others realized that some examples of homeosis in floral organs and animal skeletons could be attributed to variation in genes.
[59] The genetic studies by Morgan and others provided the foundation for the systematic analyses of Edward B. Lewis and Thomas Kaufman, which provided preliminary definitions of the many homeotic genes of the Bithorax and Antennapedia complexes, and also showed that the mutant phenotypes for most of these genes could be traced back to patterning defects in the embryonic body plan.
Hox genes play critical roles in the development of structures such as limbs, lungs, the nervous system, and eyes.
In the future, more research can be done in investigating the roles of Hox genes in leukaemia and cancer (such as EOC).