DNA replication
The double helix describes the appearance of a double-stranded DNA which is composed of two linear strands that run opposite to each other and twist together.
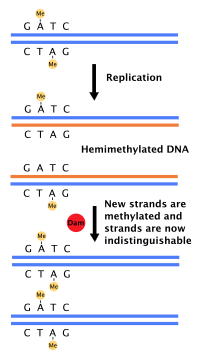
Each strand of the original DNA molecule then serves as a template for the production of its counterpart, a process referred to as semiconservative replication.
In March 2021, researchers reported evidence suggesting that a preliminary form of transfer RNA, a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code, could have been a replicator molecule itself in the very early development of life, or abiogenesis.
These nucleotides form phosphodiester bonds, creating the phosphate-deoxyribose backbone of the DNA double helix with the nucleobases pointing inward (i.e., toward the opposing strand).
[citation needed] The pairing of complementary bases in DNA (through hydrogen bonding) means that the information contained within each strand is redundant.
The actual job of the phosphodiester bonds is where in DNA polymers connect the 5' carbon atom of one nucleotide to the 3' carbon atom of another nucleotide, while the hydrogen bonds stabilize DNA double helices across the helix axis but not in the direction of the axis.
To begin synthesis, a short fragment of RNA, called a primer, must be created and paired with the template DNA strand.
[Note 1] In general, DNA polymerases are highly accurate, with an intrinsic error rate of less than one mistake for every 107 nucleotides added.
[23] Some DNA polymerases can also delete nucleotides from the end of a developing strand in order to fix mismatched bases.
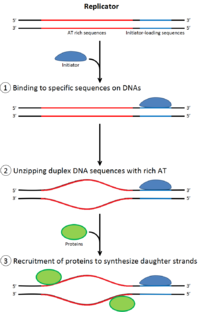
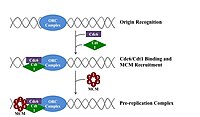
[25] DNA replication, like all biological polymerization processes, proceeds in three enzymatically catalyzed and coordinated steps: initiation, elongation and termination.
In addition, a recent report suggests that budding yeast ORC dimerizes in a cell cycle dependent manner to control licensing.
In eukaryotes, the Mcm complex is the helicase that will split the DNA helix at the replication forks and origins.
[32] If environmental conditions are right in late G1 phase, the G1 and G1/S cyclin-Cdk complexes are activated, which stimulate expression of genes that encode components of the DNA synthetic machinery.
Together, the G1/S-Cdks and/or S-Cdks and Cdc7 collaborate to directly activate the replication origins, leading to initiation of DNA synthesis.
Loading the preinitiation complex onto the origin activates the Mcm helicase, causing unwinding of the DNA helix.
Four distinct mechanisms for DNA synthesis are recognized:[citation needed] Cellular organisms use the first of these pathways since it is the most well-known.
Bacteria use a primase belonging to the DnaG protein superfamily which contains a catalytic domain of the TOPRIM fold type.
[citation needed] The primase used by archaea and eukaryotes, in contrast, contains a highly derived version of the RNA recognition motif (RRM).
[citation needed] In eukaryotes, the low-processivity enzyme, Pol α, helps to initiate replication because it forms a complex with primase.
It is produced by enzymes called helicases that break the hydrogen bonds that hold the DNA strands together in a helix.
The following is a list of major DNA replication enzymes that participate in the replisome:[46] In vitro single-molecule experiments (using optical tweezers and magnetic tweezers) have found synergetic interactions between the replisome enzymes (helicase, polymerase, and Single-strand DNA-binding protein) and with the DNA replication fork enhancing DNA-unwinding and DNA-replication.
Peter Meister et al. observed directly replication sites in budding yeast by monitoring green fluorescent protein (GFP)-tagged DNA polymerases α.
If replication forks move freely in chromosomes, catenation of nuclei is aggravated and impedes mitotic segregation.
Telomeres are regions of repetitive DNA close to the ends and help prevent loss of genes due to this shortening.
Within the germ cell line, which passes DNA to the next generation, telomerase extends the repetitive sequences of the telomere region to prevent degradation.
E. coli regulates this process through the use of termination sequences that, when bound by the Tus protein, enable only one direction of replication fork to pass through.
[32] Activation of S-Cdks in early S phase promotes the destruction or inhibition of individual pre-replication complex components, preventing immediate reassembly.
Progress of replication forks is inhibited by many factors; collision with proteins or with complexes binding strongly on DNA, deficiency of dNTPs, nicks on template DNAs and so on.
[citation needed] Most bacteria do not go through a well-defined cell cycle but instead continuously copy their DNA; during rapid growth, this can result in the concurrent occurrence of multiple rounds of replication.
[citation needed] In fast-growing bacteria, such as E. coli, chromosome replication takes more time than dividing the cell.
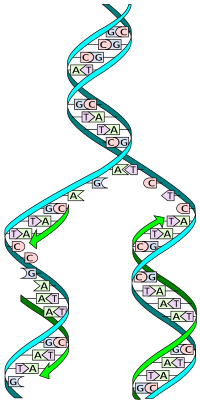
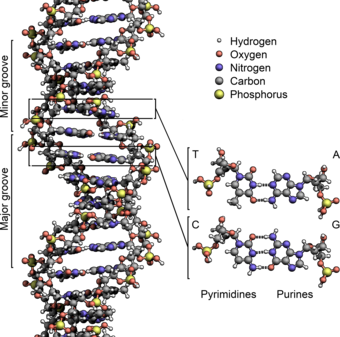


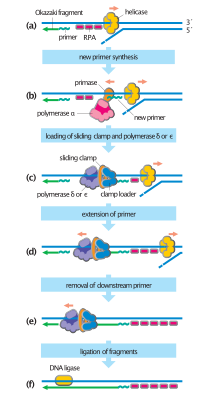
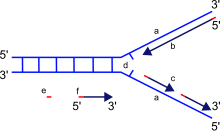
a: template, b: leading strand, c: lagging strand, d: replication fork, e: primer, f: Okazaki fragments