NetworkX
The package was crafted with the aim of creating tools to analyze data and intervention strategies for controlling the epidemic spread of disease, while also exploring the structure and dynamics of more general social, biological, and infrastructural systems.
[4] Inspired by Guido van Rossum's 1998 essay on Python graph representation,[5] NetworkX made its public debut at the 2004 SciPy annual conference.
[7] This substantial adoption rate could potentially be attributed to NetworkX's early release and its continued evolution within the SciPy ecosystem.
In 2008, SageMath, an open source mathematics system, incorporated NetworkX into its package and added support for more graphing algorithms and functions.
Visualizing large or densely connected graphs may require specialized techniques and external libraries beyond the capabilities of NetworkX alone.
These layout algorithms determine the positions of nodes and edges in a graph visualization, aiming to reveal its structure and relationships effectively.
[15] The Circular layout arranges nodes evenly around a circle, with edges drawn as straight lines connecting them.
[17] The Kamada-Kawai layout algorithm positions nodes based on their pairwise distances, aiming to minimize the total energy of the system.
It takes into account both the graph's topology and edge lengths, resulting in a layout that emphasizes geometric accuracy and readability.
[18] NetworkX provides functions for applying different layout algorithms to graphs and visualizing the results using Matplotlib or other plotting libraries.
Users can specify the desired layout algorithm when calling the drawing functions, allowing for flexible and customizable graph visualizations.
It is used widely on many levels, ranging from computer science and data analysis education to large-scale scientific studies.
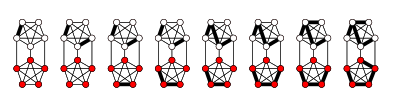
[4] The figure below demonstrates a simple example of the software's ability to create and modify variations across large amounts of networks.
In the study, NetworkX was used to find information on degree, shortest paths, clustering, and k-cores as the model introduced infections and simulated their spread.