Protein–protein interaction
Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context.
PPIs have been studied with many methods and from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal transduction, among others.
[20] In this technique the angles and intensities of a beam of X-rays diffracted by crystalline atoms are detected in a film, thus producing a three-dimensional picture of the density of electrons within the crystal.
Here are some examples of such domains: The study of the molecular structure can give fine details about the interface that enables the interaction between proteins.
[26] Based on three structures – insulin dimer, trypsin-pancreatic trypsin inhibitor complex, and oxyhaemoglobin – Cyrus Chothia and Joel Janin found that between 1,130 and 1,720 Å2 of surface area was removed from contact with water indicating that hydrophobicity is a major factor of stabilization of PPIs.
The most conventional and widely used high-throughput methods are yeast two-hybrid screening and affinity purification coupled to mass spectrometry.
[29][30] Yeast two hybrid allows the identification of pairwise PPIs (binary method) in vivo, in which the two proteins are tested for biophysically direct interaction.
The number of PPIs identified is usually low because of a high false negative rate;[34] and, understates membrane proteins, for example.
One of the most advantageous and widely used methods to purify proteins with very low contaminating background is the tandem affinity purification, developed by Bertrand Seraphin and Matthias Mann and respective colleagues.
An analysis of the results from such studies led to the conclusion that intragenic complementation, in general, arises from the interaction of differently defective polypeptide monomers to form a multimer.
Direct interaction of two nascent proteins emerging from nearby ribosomes appears to be a general mechanism for homo-oligomer (multimer) formation.
These include co-immunoprecipitation, protein microarrays, analytical ultracentrifugation, light scattering, fluorescence spectroscopy, luminescence-based mammalian interactome mapping (LUMIER), resonance-energy transfer systems, mammalian protein–protein interaction trap, electro-switchable biosurfaces, protein–fragment complementation assay, as well as real-time label-free measurements by surface plasmon resonance, and calorimetry.
The Conserved Neighborhood method is based on the hypothesis that if genes encoding two proteins are neighbors on a chromosome in many genomes, then they are likely functionally related (and possibly physically interacting).
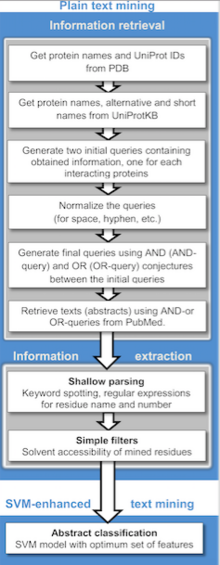
Publicly available information from biomedical documents is readily accessible through the internet and is becoming a powerful resource for collecting known protein–protein interactions (PPIs), PPI prediction and protein docking.
Currently, text mining methods generally detect binary relations between interacting proteins from individual sentences using rule/pattern-based information extraction and machine learning approaches.
There are also studies using phylogenetic profiling, basing their functionalities on the theory that proteins involved in common pathways co-evolve in a correlated fashion across species.
Some more complex text mining methodologies use advanced Natural Language Processing (NLP) techniques and build knowledge networks (for example, considering gene names as nodes and verbs as edges).
[53] Prediction models using machine learning techniques can be broadly classified into two main groups: supervised and unsupervised, based on the labeling of input variables according to the expected outcome.
[56] In 2006, random forest, an example of a supervised technique, was found to be the most-effective machine learning method for protein interaction prediction.
[59] As of 2020, a model using residue cluster classes (RCCs), constructed from the 3DID and Negatome databases, resulted in 96-99% correctly classified instances of protein–protein interactions.
[61] Large scale identification of PPIs generated hundreds of thousands of interactions, which were collected together in specialized biological databases that are continuously updated in order to provide complete interactomes.
[62] Primary databases collect information about published PPIs proven to exist via small-scale or large-scale experimental methods.
[66] However, it is important to note that some of the interactions in the STRING database are only predicted by computational methods such as Genomic Context and not experimentally verified.
Although the PPI network of a given query protein can be represented in textbooks, diagrams of whole cell PPIs are frankly complex and difficult to generate.
[68] Drawing on Kohn's map, Schwikowski et al. in 2000 published a paper on PPIs in yeast, linking 1,548 interacting proteins determined by two-hybrid screening.
[69] Bioinformatic tools have been developed to simplify the difficult task of visualizing molecular interaction networks and complement them with other types of data.
[76] Protein–protein interaction networks are often constructed as a result of lab experiments such as yeast two-hybrid screens or 'affinity purification and subsequent mass spectrometry techniques.
RNA interference (RNAi) screens (repression of individual proteins between transcription and translation) are one method that can be utilized in the process of providing signs to the protein–protein interactions.
[79][80] Nevertheless, very few PPIs are directly targeted by FDA-approved small-molecule PPI inhibitors, emphasizing a huge untapped opportunity for drug discovery.
[83] As the "modulation" of PPIs not only includes the inhibition, but also the stabilization of quaternary protein complexes, molecules with this mechanism of action (so called molecular glues) are also intensively studied.