RNA virus
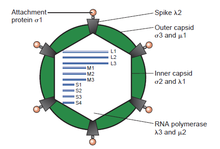
[6] The double-stranded (ds)RNA viruses represent a diverse group of viruses that vary widely in host range (humans, animals, plants, fungi,[b] and bacteria), genome segment number (one to twelve), and virion organization (Triangulation number, capsid layers, spikes, turrets, etc.).
In recent years, progress has been made in determining atomic and subnanometer resolution structures of a number of key viral proteins and virion capsids of several dsRNA viruses, highlighting the significant parallels in the structure and replicative processes of many of these viruses.
[9] The genetic diversity of RNA viruses is one reason why it is difficult to make effective vaccines against them.
For example, the region of the hepatitis C virus genome that encodes the core protein is highly conserved,[12] because it contains an RNA structure involved in an internal ribosome entry site.
[14] The sequence complexity of viruses has been shown to be a key characteristic for accurate reference-free viral classification.
After this DNA is integrated into the host genome using the viral enzyme integrase, expression of the encoded genes may lead to the formation of new virions.
Numerous RNA viruses are capable of genetic recombination when at least two viral genomes are present in the same host cell.
[16] RNA recombination appears to be a major driving force in determining genome architecture and the course of viral evolution among Picornaviridae ((+)ssRNA), e.g.
[17] In the Retroviridae ((+)ssRNA), e.g. HIV, damage in the RNA genome appears to be avoided during reverse transcription by strand switching, a form of recombination.
[21] Classification is based principally on the type of genome (double-stranded, negative- or positive-single-strand) and gene number and organization.
This is the single largest group of RNA viruses[23] and has been organized by the ICTV into the phyla Kitrinoviricota, Lenarviricota, and Pisuviricota in the kingdom Orthornavirae and realm Riboviria.
Three groups have been recognised:[25] A division of the alpha-like (Sindbis-like) supergroup on the basis of a novel domain located near the N termini of the proteins involved in viral replication has been proposed.
The evolution of the picornaviruses based on an analysis of their RNA polymerases and helicases appears to date to the divergence of eukaryotes.
[29] Their putative ancestors include the bacterial group II retroelements, the family of HtrA proteases and DNA bacteriophages.
The following four genera have been proposed for positive sense single stranded RNA satellite viruses that infect plants—Albetovirus, Aumaivirus, Papanivirus and Virtovirus.
[33] A family—Sarthroviridae which includes the genus Macronovirus—has been proposed for the positive sense single stranded RNA satellite viruses that infect arthropods.