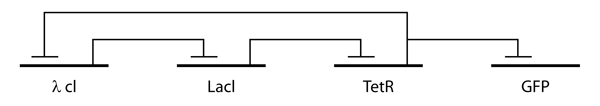
Repressilator
Artificial repressilators were first engineered by Michael Elowitz and Stanislas Leibler in 2000,[2] complementing other research projects studying simple systems of cell components and function.
In order to understand and model the design and cellular mechanisms that confers a cell’s function, Elowitz and Leibler created an artificial network consisting of a loop with three transcriptional repressors.
Six differential equations were used to model the kinetics of the repressilator system based on protein and mRNA concentrations, as well as appropriate parameter and Hill coefficient values.
It was found that these oscillations were favored by promoters coupled to efficient ribosome binding sites, cooperative transcriptional repressors, and comparable protein and mRNA decay rates.
[3] Evening-phased transcription factor CCA1 Hiking Expedition (CHE) and histone demethylase jumonji C domain-containing 5 (JMJD5) directly repress CCA1.
Other components have been found to be expressed throughout the day and either directly or indirectly inhibit or activate a consequent element in the circadian circuit, thereby creating a complex, robust and flexible network of feedback loops.
The morning phase loop has shown to be incapable of supporting circadian oscillation when evening-phase expression genes have been mutated,[5] suggesting the interdependency of each component in this naturally-occurring repressilator.
As stated in Elowitz and Leibler’s original work, the ultimate goal for repressilator research is to build an artificial circadian clock that mirrors its natural, endogenous counterpart.
This would involve developing an artificial clock with reduced noise and temperature compensation in order to better understand circadian rhythms that can be found in every domain of life.
Following noise (signal processing) analysis, the authors moved the GFP reporter construct onto the repressilator plasmid and removed the ssrA degradation tags from each repressor protein.
Further implications of this include the possibility of rescuing mutated components of oscillations synthetically in model organisms.
[7] The artificial repressilator is a milestone of synthetic biology which shows that genetic regulatory networks can be designed and implemented to perform novel functions.
New investigations at the RIKEN Quantitative Biology Center have found that chemical modifications to a single protein molecule could form a temperature independent, self-sustainable oscillator .
[13] A better understanding of the naturally-occurring repressilator in model organisms with endogenous, circadian timings, like A. thaliana, has applications in agriculture, especially in regards to plant rearing and livestock management.