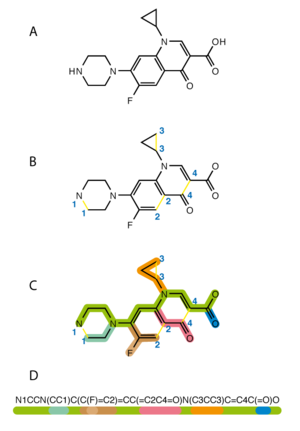
Simplified Molecular Input Line Entry System
The original SMILES specification was initiated by David Weininger at the USEPA Mid-Continent Ecology Division Laboratory in Duluth in the 1980s.
[1][2][3][4] Acknowledged for their parts in the early development were "Gilman Veith and Rose Russo (USEPA) and Albert Leo and Corwin Hansch (Pomona College) for supporting the work, and Arthur Weininger (Pomona; Daylight CIS) and Jeremy Scofield (Cedar River Software, Renton, WA) for assistance in programming the system.
In 2007, an open standard called "OpenSMILES" was developed by the Blue Obelisk open-source chemistry community.
SMILES is generally considered to have the advantage of being more human-readable than InChI; it also has a wide base of software support with extensive theoretical backing (such as graph theory).
The original paper that described the CANGEN[2] algorithm claimed to generate unique SMILES strings for graphs representing molecules, but the algorithm fails for a number of simple cases (e.g. cuneane, 1,2-dicyclopropylethane) and cannot be considered a correct method for representing a graph canonically.
In terms of a graph-based computational procedure, SMILES is a string obtained by printing the symbol nodes encountered in a depth-first tree traversal of a chemical graph.
The chemical graph is first trimmed to remove hydrogen atoms and cycles are broken to turn it into a spanning tree.
[10] Atoms are represented by the standard abbreviation of the chemical elements, in square brackets, such as [Au] for gold.
Bonds between aliphatic atoms are assumed to be single unless specified otherwise and are implied by adjacency in the SMILES string.
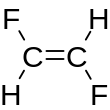
Double, triple, and quadruple bonds are represented by the symbols =, #, and $ respectively as illustrated by the SMILES O=C=O (carbon dioxide CO2), C#N (hydrogen cyanide HCN) and [Ga+]$[As-] (gallium arsenide).
For example, an alternative SMILES notation for decalin is C1CCCC2CCCCC12, where the final carbon participates in both ring-closing bonds 1 and 2.
However, they may be used with non-bonds; C1.C2.C12 is a peculiar but legal alternative way to write propane, more commonly written CCC.
Choosing a ring-break point adjacent to attached groups can lead to a simpler SMILES form by avoiding branches.
For example, cyclohexane-1,2-diol is most simply written as OC1CCCCC1O; choosing a different ring-break location produces a branched structure that requires parentheses to write.
(In fact, most SMILES software can correctly infer that the bond between the two rings cannot be aromatic and so will accept the nonstandard form c1ccccc1c2ccccc2.)
Generally, a SMILES form is easiest to read if the simpler branch comes first, with the final, unparenthesized portion being the most complex.
As a more complex example, beta-carotene has a very long backbone of alternating single and double bonds, which may be written CC1CCC/C(C)=C1/C=C/C(C)=C/C=C/C(C)=C/C=C/C=C(C)/C=C/C=C(C)/C=C/C2=C(C)/CCCC2(C)C. Configuration at tetrahedral carbon is specified by @ or @@.
While the order in which branches are specified in SMILES is normally unimportant, in this case it matters; swapping any two groups requires reversing the chirality indicator.
Other ways of writing it include C[C@H](N)C(=O)O, OC(=O)[C@@H](N)C and OC(=O)[C@H](C)N. Normally, the first of the four bonds appears to the left of the carbon atom, but if the SMILES is written beginning with the chiral carbon, such as C(C)(N)C(=O)O, then all four are to the right, but the first to appear (the [CH] bond in this case) is used as the reference to order the following three: L-alanine may also be written [C@@H](C)(N)C(=O)O.
The SMILES specification includes elaborations on the @ symbol to indicate stereochemistry around more complex chiral centers, such as trigonal bipyramidal molecular geometry.
Daylight's depict utility provides users with the means to check their own examples of SMILES and is a valuable educational tool.
While it uses many of the same symbols as SMILES, it also allows specification of wildcard atoms and bonds, which can be used to define substructural queries for chemical database searching.
One common misconception is that SMARTS-based substructural searching involves matching of SMILES and SMARTS strings.
In fact, both SMILES and SMARTS strings are first converted to internal graph representations which are searched for subgraph isomorphism.
The general syntax for the reaction extensions is REACTANT>AGENT>PRODUCT (without spaces), where any of the fields can either be left blank or filled with multiple molecules delineated with a dot (.
[15] SMILES can be converted back to two-dimensional representations using structure diagram generation (SDG) algorithms.

COc(c1)cccc1C#N
.