Transcription factor
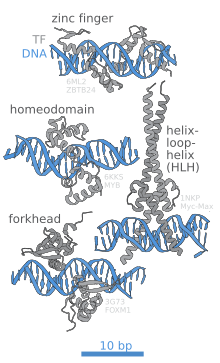
[6][7][8] A defining feature of TFs is that they contain at least one DNA-binding domain (DBD), which attaches to a specific sequence of DNA adjacent to the genes that they regulate.
[14] There are approximately 2800 proteins in the human genome that contain DNA-binding domains, and 1600 of these are presumed to function as transcription factors,[3] though other studies indicate it to be a smaller number.
[16] These mechanisms include: Transcription factors are one of the groups of proteins that read and interpret the genetic "blueprint" in the DNA.
[19][20][21] Many of these GTFs do not actually bind DNA, but rather are part of the large transcription preinitiation complex that interacts with RNA polymerase directly.
These transcription factors are critical to making sure that genes are expressed in the right cell at the right time and in the right amount, depending on the changing requirements of the organism.
The Hox transcription factor family, for example, is important for proper body pattern formation in organisms as diverse as fruit flies to humans.
[24][25] Another example is the transcription factor encoded by the sex-determining region Y (SRY) gene, which plays a major role in determining sex in humans.
Examples include heat shock factor (HSF), which upregulates genes necessary for survival at higher temperatures,[29] hypoxia inducible factor (HIF), which upregulates genes necessary for cell survival in low-oxygen environments,[30] and sterol regulatory element binding protein (SREBP), which helps maintain proper lipid levels in the cell.
[40] Transcription factors may be activated (or deactivated) through their signal-sensing domain by a number of mechanisms including: In eukaryotes, DNA is organized with the help of histones into compact particles called nucleosomes, where sequences of about 147 DNA base pairs make ~1.65 turns around histone protein octamers.
[43] Alternatively, the nucleosome can be partially unwrapped by thermal fluctuations, allowing temporary access to the transcription factor binding site.
[44] Pairs of transcription factors and other proteins can play antagonistic roles (activator versus repressor) in the regulation of the same gene.
Multiple transcription factors important in cell differentiation and lineage specification, including NANOG, SALL4A, WT1, EBF1, PU.1, and E2A, have been shown to recruit TET enzymes to specific genomic loci (primarily enhancers) to act on methylcytosine (mC) and convert it to hydroxymethylcytosine hmC (and in most cases marking them for subsequent complete demethylation to cytosine).
[62] Transcription factors interact with their binding sites using a combination of electrostatic (of which hydrogen bonds are a special case) and Van der Waals forces.
Due to the nature of these chemical interactions, most transcription factors bind DNA in a sequence specific manner.
Other constraints, such as DNA accessibility in the cell or availability of cofactors may also help dictate where a transcription factor will actually bind.
Due to their important roles in development, intercellular signaling, and cell cycle, some human diseases have been associated with mutations in transcription factors.
[64] Below are a few of the better-studied examples: Approximately 10% of currently prescribed drugs directly target the nuclear receptor class of transcription factors.
[75] Examples include tamoxifen and bicalutamide for the treatment of breast and prostate cancer, respectively, and various types of anti-inflammatory and anabolic steroids.
[77][78][79][80] Transcription factors outside the nuclear receptor family are thought to be more difficult to target with small molecule therapeutics since it is not clear that they are "drugable" but progress has been made on Pax2[81][82] and the notch pathway.
Once they occur as duplicates, accumulated mutations encoding for one copy can take place without negatively affecting the regulation of downstream targets.
However, changes of the DNA binding specificities of the single-copy Leafy transcription factor, which occurs in most land plants, have recently been elucidated.
Similar mechanisms have been proposed in the context of all alternative phylogenetic hypotheses, and the role of transcription factors in the evolution of all species.
The resistant to oxidative stress and alkaline pH sensing were contributed from the transcription factor Yap1 and Rim101 of the Papiliotrema terrestris LS28 as molecular tools revealed an understanding of the genetic mechanisms underlying the biocontrol activity which supports disease management programs based on biological and integrated control.
[citation needed] The most commonly used method for identifying transcription factor binding sites is chromatin immunoprecipitation (ChIP).
[89] This technique relies on chemical fixation of chromatin with formaldehyde, followed by co-precipitation of DNA and the transcription factor of interest using an antibody that specifically targets that protein.
[90] As described in more detail below, transcription factors may be classified by their (1) mechanism of action, (2) regulatory function, or (3) sequence homology (and hence structural similarity) in their DNA-binding domains.