Twitching motility
[1][2][3] The name twitching motility is derived from the characteristic jerky and irregular motions of individual cells when viewed under the microscope.
Active movement mediated by the twitching system has been shown to be an important component of the pathogenic mechanisms of several species.
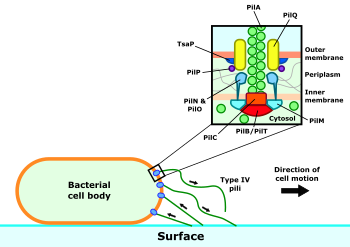
Surrounding the motor complex is the alignment subcomplex, formed from the PilM, PilN, PilO and PilP proteins.
[9] Regulatory proteins associated with the twitching motility system have strong sequence and structural similarity to those that regulate bacterial chemotaxis using flagellae.
[18] Both presence of type IV pili and active pilar movement appear to be important contributors to the pathogenicity of several species.
[8] In P. aeruginosa, loss of pilus retraction results in a reduction of bacterial virulence in pneumonia[19] and reduces colonisation of the cornea.
[20] Some bacteria are also able to twitch along vessel walls against the direction of fluid flow within them,[21] which is thought to permit colonisation of otherwise inaccessible sites in the vasculatures of plants and animals.
This positive feedback is an important initiating factor for the establishment of microcolonies, the precursors to fully fledged biofilms.
[23][26] Type IV pili and related structures can be found across almost all phyla of Bacteria and Archaea,[27] however definitive twitching motility has been shown in a more limited range of prokaryotes.
Most well studied and wide spread are the twitching Pseudomonadota, such as Neisseria gonorrhoeae, Myxococcus xanthus and Pseudomonas aeruginosa.