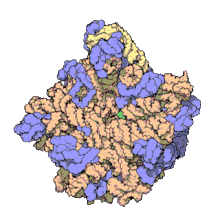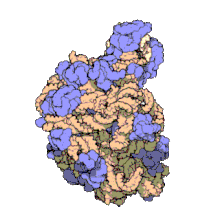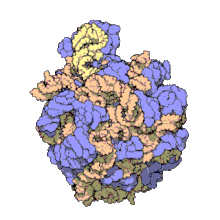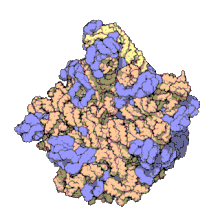
5S ribosomal RNA
[14][17] Once transcribed, the 3' ends of 5S rRNA can only be trimmed to mature length by functional homologues of RNase T, for example Rex1p in Saccharomyces cerevisiae.
Interaction of 5S rRNA with the La protein prevents the RNA from degradation by exonucleases in the cell.
[19] La protein is found in the nucleus in all eukaryotic organisms and associates with several types of RNAs transcribed by RNA pol III.
La protein interacts with these RNAs (including the 5S rRNA) through their 3' oligo-uridine tract, aiding stability and folding of the RNA.
[4][20] In eukaryotic cells, ribosomal protein L5 associates and stabilizes the 5S rRNA forming a pre-ribosomal ribonucleoprotein particle (RNP) that is found in both cytosol and the nucleus.
L5 deficiency prevents transport of 5S rRNA to the nucleus and results in decreased ribosomal assembly.
Organelle genomes encode SSU and LSU rRNAs without exception, yet the distribution of 5S rRNA genes (rrn5) is most uneven.
[22][23] Additional, more divergent organellar 5S rRNAs were only identified with specialized covariance models that incorporate information on the pronounced sequence composition bias and structural variation.
Current evidence indicates that mitochondrial DNA of only a few groups, notably animals, fungi, alveolates and euglenozoans lacks the gene.
[24] The central protuberance, otherwise occupied by 5S rRNA and its associated proteins (see Figure 2), was remodeled in various ways.