Sense (molecular biology)
Because of the complementary nature of base-pairing between nucleic acid polymers, a double-stranded DNA molecule will be composed of two strands with sequences that are reverse complements of each other.
The terms "sense" and "antisense" are relative only to the particular RNA transcript in question, and not to the DNA strand as a whole.
For example, the sequence "ATG" within a DNA sense strand corresponds to an "AUG" codon in the mRNA, which codes for the amino acid methionine.
The only biological information that is important for labeling strands is the relative locations of the terminal 5′ phosphate group and the terminal 3′ hydroxyl group (at the ends of the strand or sequence in question), because these ends determine the direction of transcription and translation.
For example, the notation "YEL021W", an alias of the URA3 gene used in the National Center for Biotechnology Information (NCBI) database, denotes that this gene is in the 21st open reading frame (ORF) from the centromere of the left arm (L) of Yeast (Y) chromosome number V (E), and that the expression coding strand is the Watson strand (W).
Bunyaviruses have three single-stranded RNA (ssRNA) fragments, some of them containing both positive-sense and negative-sense sections; arenaviruses are also ssRNA viruses with an ambisense genome, as they have three fragments that are mainly negative-sense except for part of the 5′ ends of the large and small segments of their genome.
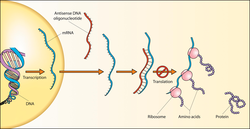
The concept has also been exploited as a molecular biology technique, by artificially introducing a transgene coding for antisense RNA in order to block the expression of a gene of interest.
Radioactively or fluorescently labelled antisense RNA can be used to show the level of transcription of genes in various cell types.
In the United States, the Food and Drug Administration (FDA) has approved the phosphorothioate antisense oligonucleotides fomivirsen (Vitravene)[5] and mipomersen (Kynamro)[6] for human therapeutic use.
Some viruses (e.g. Coronaviridae) have positive-sense genomes that can act as mRNA and be used directly to synthesize proteins without the help of a complementary RNA intermediate.
Gene silencing can be achieved by introducing into cells a short "antisense oligonucleotide" that is complementary to an RNA target.
Double-stranded RNA can also act as a catalytic, enzyme-dependent antisense agent through the RNAi/siRNA pathway, involving target mRNA recognition through sense-antisense strand pairing followed by target mRNA degradation by the RNA-induced silencing complex (RISC).
The R1 plasmid hok/sok system provides yet another example of an enzyme-dependent antisense regulation process through enzymatic degradation of the resulting RNA duplex.
Other antisense mechanisms are not enzyme-dependent, but involve steric blocking of their target RNA (e.g. to prevent translation or to induce alternative splicing).