Bacterial taxonomy
This name denotes the two lowest levels in a hierarchy of ranks, increasingly larger groupings of species based on common traits.
[4][5][6] Early described genera of bacteria include Vibrio and Monas, by O. F. Müller (1773, 1786), then classified as Infusoria (however, many species before included in those genera are regarded today as protists); Polyangium, by H. F. Link (1809), the first bacterium still recognized today; Serratia, by Bizio (1823); and Spirillum, Spirochaeta and Bacterium, by Ehrenberg (1838).
[7] Bacteria were first classified as plants constituting the class Schizomycetes, which along with the Schizophyceae (blue green algae/Cyanobacteria) formed the phylum Schizophyta.
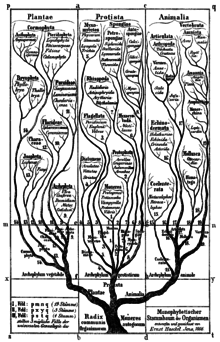
[11] Haeckel in 1866 placed the group in the phylum Moneres (from μονήρης: simple) in the kingdom Protista and defines them as completely structureless and homogeneous organisms, consisting only of a piece of plasma.
[10] He subdivided the phylum into two groups:[10] The classification of Ferdinand Cohn (1872) was influential in the nineteenth century, and recognized six genera: Micrococcus, Bacterium, Bacillus, Vibrio, Spirillum, and Spirochaeta.
in 1905, Erwin F. Smith accepted 33 valid different names of bacterial genera and over 150 invalid names,[17] and Vuillemin, in a 1913 study,[18] concluded that all species of the Bacteria should fall into the genera Planococcus, Streptococcus, Klebsiella, Merista, Planomerista, Neisseria, Sarcina, Planosarcina, Metabacterium, Clostridium, Serratia, Bacterium, and Spirillum.
Bergey et al.[24] presented a classification which generally followed the 1920 Final Report of the Society of American Bacteriologists Committee (Winslow et al.), which divided class Schizomycetes into four orders: Myxobacteriales, Thiobacteriales, Chlamydobacteriales, and Eubacteriales, with a fifth group being four genera considered intermediate between bacteria and protozoans: Spirocheta, Cristospira, Saprospira, and Treponema.
However, different authors often reclassified the genera due to the lack of visible traits to go by, resulting in a poor state which was summarised in 1915 by Robert Earle Buchanan.
[30] A. R. Prévot's system[31][32]) had four subphyla and eight classes, as follows: Despite there being little agreement on the major subgroups of the Bacteria, Gram staining results were most commonly used as a classification tool.
Consequently, until the advent of molecular phylogeny, the Kingdom Prokaryota was divided into four divisions,[41] A classification scheme still formally followed by Bergey's manual of systematic bacteriology for tome order[42] Woese argued that the bacteria, archaea, and eukaryotes represent separate lines of descent that diverged early on from an ancestral colony of organisms.
[47] In any case, it is thought that viruses and archaea began relationships approximately two billion years ago, and that co-evolution may have been occurring between members of these groups.
[50] With improved methodologies it became clear that the methanogenic bacteria were profoundly different and were (erroneously) believed to be relics of ancient bacteria[51] thus Carl Woese, regarded as the forerunner of the molecular phylogeny revolution, identified three primary lines of descent: the Archaebacteria, the Eubacteria, and the Urkaryotes, the latter now represented by the nucleocytoplasmic component of the Eukaryotes.
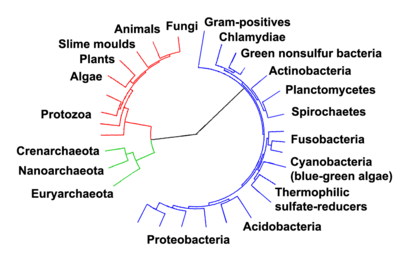
[53] In 1987 Carl Woese divided the Eubacteria into 11 divisions based on 16S ribosomal RNA (SSU) sequences, which with several additions are still used today.
[54][55] Oren and Goker has also validly published a number of kingdoms as a layer higher than the division/phylum:[53] While the three domain system is widely accepted,[56] some authors have opposed it for various reasons.
One prominent scientist who opposes the three domain system is Thomas Cavalier-Smith, who proposed that the Archaea and the Eukaryotes (the Neomura) stem from Gram positive bacteria (Posibacteria), which in turn derive from gram negative bacteria (Negibacteria) based on several logical arguments,[57][58] which are highly controversial and generally disregarded by the molecular biology community (c.f.
[60] However, despite there being a wealth of statistically supported studies towards the rooting of the tree of life between the Bacteria and the Neomura by means of a variety of methods,[61] including some that are impervious to accelerated evolution—which is claimed by Cavalier-Smith to be the source of the supposed fallacy in molecular methods[57]—there are a few studies which have drawn different conclusions, some of which place the root in the phylum Firmicutes with nested archaea.
[68] The only cladistic analyses for bacteria based on classical evidence largely corroborate Gupta's results (see comprehensive mega-taxonomy).
James Lake presented a 2 primary kingdom arrangement (Parkaryotae + eukaryotes and eocytes + Karyotae) and suggested a 5 primary kingdom scheme (Eukaryota, Eocyta, Methanobacteria, Halobacteria, and Eubacteria) based on ribosomal structure and a 4 primary kingdom scheme (Eukaryota, Eocyta, Methanobacteria, and Photocyta), bacteria being classified according to 3 major biochemical innovations: photosynthesis (Photocyta), methanogenesis (Methanobacteria), and sulfur respiration (Eocyta).
[75]: 63 Today the taxa which have been correctly described are reviewed in Bergey's manual of Systematic Bacteriology, which aims to aid in the identification of species and is considered the highest authority.
e.g. "Candidatus Pelagibacter ubique" Bacteria divide asexually and for the most part do not show regionalisms ("Everything is everywhere"), therefore the concept of species, which works best for animals, becomes entirely a matter of judgment.
The number of named species of bacteria and archaea (approximately 21,000)[88] is surprisingly small considering their early evolution, genetic diversity and residence in all ecosystems.
[90] However, a quicker diagnostic ad hoc threshold to separate species is less than 70% DNA–DNA hybridisation,[91] which corresponds to less than 97% 16S DNA sequence identity.
In the family Enterobacteriaceae of the class Gammaproteobacteria, the species in the genus Shigella (S. dysenteriae, S. flexneri, S. boydii, S. sonnei) from an evolutionary point of view are strains of the species Escherichia coli (polyphyletic), but due to genetic differences cause different medical conditions in the case of the pathogenic strains.
thuringiensis, B. mycoides, B. pseudomycoides, B. weihenstephanensis and B. medusa) have 99-100% similar 16S rRNA sequence (97% is a commonly cited adequate species cut-off) and are polyphyletic, but for medical reasons (anthrax etc.)
nov.: Rhizobium radiobacter (formerly known as A. tumefaciens), R. rhizogenes, R. rubi, R. undicola and R. vitis)[102] Given the plant pathogenic nature of Agrobacterium species, it was proposed to maintain the genus Agrobacterium[103] and the latter was counter-argued[104] Taxonomic names are written in italics (or underlined when handwritten) with a majuscule first letter with the exception of epithets for species and subspecies.
The category of kingdom was included into the Bacteriological Code in November 2023,[116] the first four proposals (Bacillati, Fusobacteriati, Pseudomonadati, Thermotogati) were validly published in January 2024.
Some unusual bacteria and archaea have special names such as Quin's oval (Quinella ovalis) and Walsby's square (Haloquadratum walsbyi).