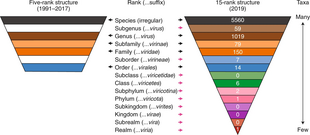
Virus classification
In 2021, the ICTV changed the International Code of Virus Classification and Nomenclature (ICVCN) to mandate a binomial format (genus|| ||species) for naming new viral species similar to that used for cellular organisms; the names of species coined prior to 2021 are gradually being converted to the new format, a process planned for completion by the end of 2023.
The currently accepted and formal definition of a 'virus' was accepted by the ICTV Executive Committee in November 2020 and ratified in March 2021, and is as follows:[2] Viruses sensu stricto are defined operationally by the ICTV as a type of MGE that encodes at least one protein that is a major component of the virion encasing the nucleic acid of the respective MGE and therefore the gene encoding the major virion protein itself or MGEs that are clearly demonstrable to be members of a line of evolutionary descent of such major virion protein-encoding entities.
Before 1982, it was thought that viruses could not be made to fit Ernst Mayr's reproductive concept of species, and so were not amenable to such treatment.
These criteria may include, but are not limited to, natural and experimental host range, cell and tissue tropism, pathogenicity, vector specificity, antigenicity, and the degree of relatedness of their genomes or genes.
The criteria used should be published in the relevant section of the ICTV Report and reviewed periodically by the appropriate Study Group.
[10] Therefore, structural relationship between viruses has been suggested to be used as a basis for defining higher-level taxa – structure-based viral lineages – that could complement the ICTV classification scheme of 2010.
[13] Named after David Baltimore, a Nobel Prize-winning biologist, these groups are designated by Roman numerals.
Classifying viruses according to their genome means that those in a given category will all behave in a similar fashion, offering some indication of how to proceed with further research.
[16] All viruses that encode a reverse transcriptase (also known as RT or RNA-dependent DNA polymerase) are members of the class Revtraviricetes, within the phylum Arterviricota, kingdom Pararnavirae, and realm Riboviria.
[17] Holmes (1948) used a Linnaean taxonomy with binomial nomenclature to classify viruses into 3 groups under one order, Virales.
The prototypical example is members of the (also polyphyletic) Polydnaviriformidae, which are used by wasps to send pieces of immunity-blunting DNA into the prey by packing them into virion-like particles.
GTA particles resemble tailed phages, but are smaller and carry mostly random pieces of host DNA.
[25] The three known clades of GTAs, Rhodogtaviriformidae, Bartogtaviriformidae, and Brachygtaviriformidae, all arose independently from different parts of the Caudoviricetes family tree.