Betalipothrixvirus
[1][2] All species within the genus have linear double-stranded DNA genomes and some infect hyperthermophilic crenarchaea of the orders Sulfolobales.
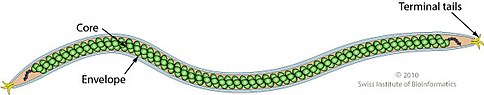
[3] Etymology: the root of the word (Beta)Lipothrix: from Greek, "fat hair", referring to the virion shape(SIB).
For example, the core of the SIFV species is formed by “a zipper-like array of DNA-associated protein subunits and is covered by a lipid envelope containing host lipids.”[4] Similarly, in images of the structure of AFV3, zipper or zigzag patterned high-density “ovoid masses” have been found inside the lumen of the virus.
These large regions have been predicted to produce a putative helicase, a nuclease, a protein phosphatase, transcription regulators and glycosyltransferases.
[6] Furthermore, studies have shown that some of the shared features of the genomes of these six species include (1) large inverted terminal repeats exhibiting conserved, regularly spaced direct repeats; (2) a highly conserved operon encoding the two major structural proteins; (3) multiple overlapping open reading frames, which may be indicative of gene recoding; (4) putative 12-bp genetic elements; and (5) partial gene sequences corresponding closely to spacer sequences of chromosomal repeat clusters.