BioBIKE
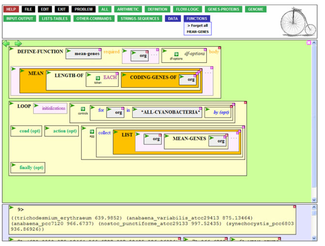
[4] BioBIKE is an integrated symbolic biocomputing and bioinformatics platform, built from the start as an entirely (what is now called) cloud-based architecture where all computing is done in remote servers, and all user access is accomplished through web browsers.
This enables code written either in the native Lisp, in the visual programming language, or systems of rules expressed in the SNARK theorem prover to access the whole of biological knowledge in an integrated manner.
For its time (released in 2002) it was unique in permitting users to create fully functional biocomputing programs that run on the back-end servers entirely through the web browser UI.
[1] Being a multi-headed, multi-threaded, multi-user, multi-tenancy cloud-based system, BioBIKE users were able to directly work together through their web browsers, remotely sharing the same listener and memory space.
[5] A specialized offshoot of BioBIKE called "BioDeducta" includes SRI's SNARK theorem prover, offering unique "deductive biocomputing" capabilities.