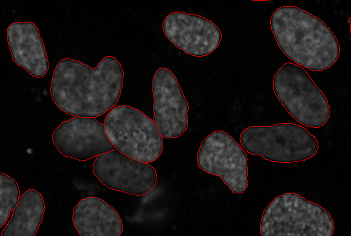
Bioimage informatics
[1] It focuses on the use of computational techniques to analyze bioimages, especially cellular and molecular images, at large scale and high throughput.
[2][3] In addition, automated systems are unbiased, unlike human based analysis whose evaluation may (even unconsciously) be influenced by the desired outcome.
There has been an increasing focus on developing novel image processing, computer vision, data mining, database and visualization techniques to extract, compare, search and manage the biological knowledge in these data-intensive problems.
Confocal stacks will require 3D processing and widefield pseudo-stacks will often benefit from digital deconvolution to remove the out-of-focus light.
The advent of automated microscopes that can acquire many images automatically is one of the reasons why analysis cannot be done by eye (otherwise, annotation would rapidly become the research bottleneck).
On the other hand, the operator might introduce an unconscious bias in his selection by choosing only the cells whose phenotype is most like the one expected before the experiment.
If the goal is a medical diagnostic, then histology applications will often fall into the realm of digital pathology or automated tissue image analysis, which are sister fields of bioimage informatics.
Using multi-well plates, robotics, and automated microscopy, the same assay can be applied to a large library of possible reagents (typically either small molecules or RNAi) very rapidly, obtaining thousands of images in a short amount of time.
As obtaining high contrast is done by shining more light which damages the sample and destroys the dye, illumination is kept at a minimum.
One example is as time-course data is collected, images in subsequent frames must often be registered so that minor shifts in the camera position can be corrected for.
It has been primarily used to generate more than 50,000 3D standardized fruitfly brain images at Janelia Farm of HHMI, with other applications including dragonfly and mice.
The OpenAccess journal BMC Bioinformatics has a section devoted to bioimage analysis, visualization and related applications.
There are several packages that make bioimage informatics methods available through a graphical user interface such as ImageJ, FIJI, CellProfiler, chunkflow or Icy.
Visualization and analysis platforms such as Vaa3D have appeared in recent years and have been used in both large scale projects especially for neuroscience and desktop applications.
Other researchers develop their own methods, typically based on a programming language with good computer vision support such as Python, C++, or MATLAB.
Although, examples of researcher developed methods in programming languages with less computer vision support as R exist (e.g. trackdem [14]).