SARS-CoV-2
This was in accordance with WHO's 2015 guidance[30] against using geographical locations, animal species, or groups of people in disease and virus names.
[31][32] On 11 February 2020, the International Committee on Taxonomy of Viruses adopted the official name "severe acute respiratory syndrome coronavirus 2" (SARS‑CoV‑2).
[33] To avoid confusion with the disease SARS, the WHO sometimes refers to SARS‑CoV‑2 as "the COVID-19 virus" in public health communications[34][35] and the name HCoV-19 was included in some research articles.
[16][39][40][41] Transmission was initially assumed to occur primarily via respiratory droplets from coughs and sneezes within a range of about 1.8 metres (6 ft).
[42][43] Laser light scattering experiments suggest that speaking is an additional mode of transmission[44][45] and a far-reaching[46] one, indoors, with little air flow.
[59][60] The degree to which the virus is infectious during the incubation period is uncertain, but research has indicated that the pharynx reaches peak viral load approximately four days after infection[61][62] or in the first week of symptoms and declines thereafter.
[64] A study by a team of researchers from the University of North Carolina found that the nasal cavity is seemingly the dominant initial site of infection, with subsequent aspiration-mediated virus-seeding into the lungs in SARS‑CoV‑2 pathogenesis.
[65] Studies have identified a range of animals—such as cats, ferrets, hamsters, non-human primates, minks, tree shrews, raccoon dogs, fruit bats, and rabbits—that are susceptible and permissive to SARS-CoV-2 infection.
[73] That may explain how out of 217 on board a cruise liner that docked at Montevideo, only 24 of 128 who tested positive for viral RNA showed symptoms.
[53] The authors later published a correction that showed that shedding began earlier than first estimated, four to five days before symptoms appear.
Since genomic analyses showed significant genetic differences between the SARS‑CoV‑2 variant sampled on those two dates, the case study authors determined this was a reinfection.
[79] Prior to the emergence of SARS-CoV-2 as a pathogen infecting humans, there had been two previous zoonosis-based coronavirus epidemics, those caused by SARS-CoV-1 and MERS-CoV.
[80] The original source of viral transmission to humans remains unclear, as does whether the virus became pathogenic before or after the spillover event.
[86] Coronaviruses in general have high genetic plasticity,[89] but SARS-CoV-2's viral evolution is slowed by the RNA proofreading capability of its replication machinery.
The closest match by far, published in Nature (journal) in February 2022, were viruses BANAL-52 (96.8% resemblance to SARS‑CoV‑2), BANAL-103 and BANAL-236, collected in three different species of bats in Feuang, Laos.
[92][93][94] An earlier source published in February 2020 identified the virus RaTG13, collected in bats in Mojiang, Yunnan, China to be the closest to SARS‑CoV‑2, with 96.1% resemblance.
In addition, despite similarities in a few critical amino acids,[102] pangolin virus samples exhibit poor binding to the human ACE2 receptor.
[104] Human coronaviruses are capable of causing illnesses ranging from the common cold to more severe diseases such as Middle East respiratory syndrome (MERS, fatality rate ~34%).
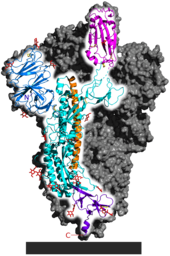
[108] A distinguishing feature of SARS‑CoV‑2 is its incorporation of a polybasic site cleaved by furin,[102][112] which appears to be an important element enhancing its virulence.
[114] The furin protease recognizes the canonical peptide sequence RX[R/K] R↓X where the cleavage site is indicated by a down arrow and X is any amino acid.
[120] Viral genetic sequence data can provide critical information about whether viruses separated by time and space are likely to be epidemiologically linked.
[121] With a sufficient number of sequenced genomes, it is possible to reconstruct a phylogenetic tree of the mutation history of a family of viruses.
[123] Examination of the topology of the phylogenetic tree at the start of the pandemic also found high similarities between human isolates.
[125] On 11 February 2020, the International Committee on Taxonomy of Viruses announced that according to existing rules that compute hierarchical relationships among coronaviruses based on five conserved sequences of nucleic acids, the differences between what was then called 2019-nCoV and the virus from the 2003 SARS outbreak were insufficient to make them separate viral species.
[155] The mutation of CG dinucleotides is thought to arise to avoid the zinc finger antiviral protein related defense mechanism of cells,[156] and to lower the energy to unbind the genome during replication and translation (adenosine and uracil base pair via two hydrogen bonds, cytosine and guanine via three).
[166] SARS‑CoV‑2 produces at least three virulence factors that promote shedding of new virions from host cells and inhibit immune response.
Masitinib was found to inhibit SARS-CoV-2 main protease, showing a greater than 200-fold reduction in viral titers in the lungs and nose of mice, however it is not approved for the treatment of COVID-19 in humans.
[173] COVID Moonshot is an international collaborative open-science project started in March 2020 with the goal of developing an un-patented oral antiviral drug for treatment of SARS-CoV-2.
[174] Retrospective tests collected within the Chinese surveillance system revealed no clear indication of substantial unrecognized circulation of SARS‑CoV‑2 in Wuhan during the latter part of 2019.
For instance, one study found relatively low R0 (~3.5) in Sweden, Belgium and the Netherlands, while Spain and the US had significantly higher R0 values (5.9 to 6.4, respectively).