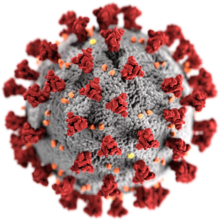
SARS-related coronavirus
Betacoronavirus pandemicum[2] (also known as Severe acute respiratory syndrome–related coronavirus, abbreviated as SARSr-CoV or SARS-CoV)[note 1] is a species of virus consisting of many known strains.
[3][5][6][7] These enveloped, positive-sense single-stranded RNA viruses enter host cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor.
[11][12] SARS-related coronavirus is a member of the genus Betacoronavirus (group 2) and monotypic of the subgenus Sarbecovirus (subgroup B).
[13] Sarbecoviruses, unlike embecoviruses or alphacoronaviruses, have only one papain-like proteinase (PLpro) instead of two in the open reading frame ORF1ab.
[14] SARSr-CoV was determined to be an early split-off from the betacoronaviruses based on a set of conserved domains that it shares with the group.
[19] The 5' methylated cap and 3' polyadenylated tail allows the positive-sense RNA genome to be directly translated by the host cell's ribosome on viral entry.
[21] SARSr-CoV is similar to other coronaviruses in that its genome expression starts with translation by the host cell's ribosomes of its initial two large overlapping open reading frames (ORFs), 1a and 1b, both of which produce polyproteins.
The spike protein's interaction with its complement host cell receptor is central in determining the tissue tropism, infectivity, and species range of the virus.
The path the virus takes depends on the host protease available to cleave and activate the receptor-attached spike protein.
[60] The attachment of sarbecoviruses to ACE2 has been shown to be an evolutionarily conserved feature, present in many species of the taxon.
[61] The first path the SARS coronavirus can take to enter the host cell is by endocytosis and uptake of the virus in an endosome.
[62][63] In the SARS coronavirus, the activation of the C-terminal part of the spike protein triggers the fusion of the viral envelope with the host cell membrane by inducing conformational changes which are not fully understood.
The larger polyprotein pp1ab is a result of a -1 ribosomal frameshift caused by a slippery sequence (UUUAAAC) and a downstream RNA pseudoknot at the end of open reading frame ORF1a.
"[69] A number of the nonstructural replication proteins coalesce to form a multi-protein replicase-transcriptase complex (RTC).
Similarly, proteins nsp7 and nsp8 form a hexadecameric sliding clamp as part of the complex which greatly increases the processivity of the RNA-dependent RNA polymerase.
[73] Human SARS-CoV appears to have had a complex history of recombination between ancestral coronaviruses that were hosted in several different animal groups.