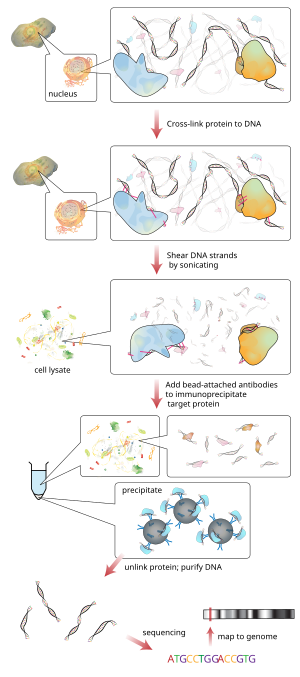
Chromatin immunoprecipitation
[2] Briefly, the conventional method is as follows: There are mainly two types of ChIP, primarily differing in the starting chromatin preparation.
Cell debris in the sheared lysate is then cleared by sedimentation and protein–DNA complexes are selectively immunoprecipitated using specific antibodies to the protein(s) of interest.
Chromatin Immunoprecipitation sequencing, also known as ChIP-seq, is an experimental technique used to identify transcription factor binding events throughout an entire genome.
ChIP-seq is the primary technique to complete this task, as it has proven to be extremely effective in resolving how proteins and transcription factors influence phenotypical mechanisms.
ChIP-on-chip, also known as ChIP-chip, is an experimental technique used to isolate and identify genomic sites occupied by specific DNA-binding proteins in living cells.
The two methods seek similar results, as they both strive to find protein binding sites that can help identify elements in the human genome.
The main difference comes from the efficacy of the two techniques, ChIP-seq produces results with higher sensitivity and spatial resolution because of the wide range of genomic coverage.
[2] Table 1 Advantages and disadvantages of NChIP and XChIP Better chromatin and protein revery efficiency due to better antibody specificity In 1984 John T. Lis and David Gilmour, at the time a graduate student in the Lis lab, used UV irradiation, a zero-length protein-nucleic acid crosslinking agent, to covalently cross-link proteins bound to DNA in living bacterial cells.
A year later they used the same methodology to study the distribution of eukaryotic RNA polymerase II on fruit fly heat shock genes.
[9][10] XChIP was further modified and developed by Alexander Varshavsky and co-workers, who examined the distribution of histone H4 on heat shock genes using formaldehyde cross-linking.