Circulating tumor DNA
[1][2] Recent studies have laid the foundation for inferring gene expression from cfDNA (and ctDNA), with EPIC-seq emerging as a notable advancement.
[3] This method has substantially raised the bar for the noninvasive inference of expression levels of individual genes, thereby augmenting the assay's applicability in disease characterization, histological classification, and monitoring treatment efficacy.
[7][8][9][10][11] Studies in both human (healthy and cancer patients)[12] and xenografted mice[13] show that the size of fragmented cfDNA is predominantly 166bp long, which corresponds to the length of DNA wrapped around a nucleosome plus a linker.
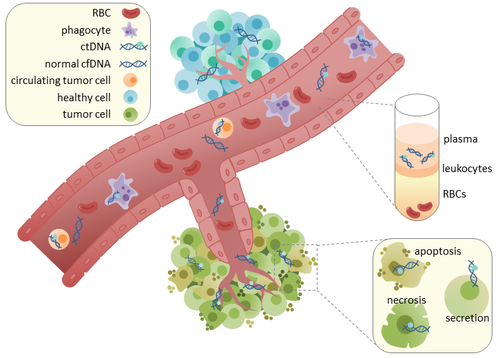
[14][15] In healthy tissue, infiltrating phagocytes are responsible for clearance of apoptotic or necrotic cellular debris, which includes cfDNA.
[17] This possibly occurs due to inefficient immune cell infiltration to tumor sites, which reduces effective clearance of ctDNA from the bloodstream.
[16] Comparison of mutations in ctDNA and DNA extracted from primary tumors of the same patients revealed the presence of identical cancer-relevant genetic changes.
[23] Sherwood et al. demonstrated superior detection of KRAS mutations in matched samples collected in both EDTA K3 and Streck BCT tubes.
[citation needed] A whole genome or whole exome sequencing approaches may be necessary to discover new mutations in tumor DNA while monitoring disease burden or tracking drug resistance.
This makes it difficult to detect rare mutations, or in situations where low ctDNA levels are present (such as minimal residual disease).
Limiting the sequencing to only the whole exome instead can decrease expense and increase speed, but at the cost of losing information about mutations in the non-coding regulatory regions of DNA.
These unique patterns can be an important source of information to improve the detection of ctDNA or localize the tissue of origin of these fragments.
[28] Size-selection of short fragments (<150bp) with in vitro or in silico methods could improve the recovery of mutations and copy number aberrations.
[15] This method was originally developed by the laboratory of Bert Vogelstein, Luis Diaz, and Victor Velculescu at Johns Hopkins University.
DNA hydroxymethylation is a similarly associated mark that has been shown to be a predictive marker of healthy versus diseased conditions in cfDNA, including cancer.
[32][33]) In a targeted approach, sequencing of ctDNA can be directed towards a genetic panel constructed based on mutational hotspots for the cancer of interest.
Then individual polymerase chain reactions occur in each droplet using selected primers against regions of ctDNA and proceeds to endpoint.
ddPCR allows for highly quantitative assessment of allele and mutant frequencies in ctDNA but is limited by the number of fluorescent probes that can be used in one assay (up to 5).
[34] Specificity should be augmented through the use of either minor groove binding (MGB) modified probes or of an alternative such as locked nucleic acids (LNAs).
Beads, emulsification, amplification, and magnetics (BEAMing) is a technique that builds upon Droplet Digital PCR in order to identify mutations in ctDNA using flow cytometry.
However, unlike with ddPCR, a larger number of DNA sequences can be interrogated due to the flexibility of using fluorescently bound probes.
[34] Cancer personalized profiling by deep sequencing (CAPP-Seq) was originally described by Ash Alizadeh and Maximilian Diehn's groups at Stanford University.
Then, a microfluidics system is used to attached adaptors with a unique identifier to each amplicon to further amplify the DNA in parallel singleplex reactions.
This technique was shown to successfully identify mutations scattered in the TP53 tumor suppressor gene in advanced ovarian cancer patients.
Safe-Seq decreases the error rate of massively parallel sequencing in order to increase the sensitivity to rare mutants.
This novel and promising technique has provided information on resistance to treatment with androgen receptor signaling inhibitors, intratumoral heterogeneity (thanks to phylogenetic evolution and molecular chronology), chromosomal instability, contribution of ctDNA to metastasis through global transcriptomic patterns (taking into account nucleosomes present in transcription start sites (TSSs) and AR-binding sites (ARBs).
This subclonal reconstruction based on ctDNA thanks to Whole-genome sequencing poses a unique set of challenges and opportunities for scientific research in oncology.
[27] However, the authors acknowledge that ctDNA analysis is not without limitations; plasma samples collected post-operatively were only able to predict recurrence at 36 months in 48% of cases.
The use of ctDNA in research can alleviate these concerns because it could provide a more representative 'screenshot' of the genetic diversity of cancer at both primary and metastatic sites.
[47] Early detection of cancer is still challenging but recent progress in the analysis of the epigenetic features of cfDNA, or the fragmentation pattern unlock improve the sensitivity of liquid biopsy.
[28] Furthermore, ctDNA analysis is an emerging tool for understanding the clonal composition of metastatic tumors, detecting different mutations on a genomic scale, studying subclonal diversity that affects the prognosis of the disease as different resistant phenotypes can be found and the appearance of new mechanisms of genomic and transcriptomic resistance to treatment.