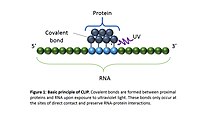
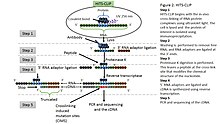
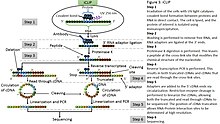
Cross-linking immunoprecipitation
[6] cDNA is then synthesized via RT-PCR followed by high-throughput sequencing followed by mapping the reads back to the transcriptome and other computational analyses to study the interaction sites.
[2] CLIP was originally undertaken to study interactions between the neuron-specific RNA-binding protein and splicing factors NOVA1 and NOVA2 in the mouse brain, identifying RNA binding sites that contained the expected Nova-binding motifs.
Sequencing of the cDNA library identified many positions close to alternative exons, several of which were found to require Nova1/2 for their brains-specific splicing patterns.
[5] HITS-CLIP also introduced the addition of dinucleotide barcodes to primers, providing the ability to sequence and then deconvolute multiple experiments simultaneously.
[28] PAR-CLIP (photoactivatable ribonucleoside–enhanced cross-linking and immunoprecipitation) is also used for identifying the binding sites of cellular RNA-binding proteins (RBPs) and microRNA-containing ribonucleoprotein complexes (miRNPs).
Irradiation of the cells by UV light of 365 nm induces efficient cross-linking of photoreactive nucleoside-labeled cellular RNAs to interacting RBPs.
[21]iCLIP (individual nucleotide–resolution crosslinking and immunoprecipitation) is a variant of CLIP that enabled amplification of truncated cDNAs, which are produced when reverse transcription stops prematurely at the cross-link site.
Additionally, the amplification in eCLIP is now comparable to RNA-seq, enabling rigorous quantitative normalization against paired input controls (to remove background at ribosomal and other highly abundant RNAs) as well as quantitative comparison across peaks and samples, enabling the ability to detect allele-specific binding or differential RNA binding between conditions.As in other CLIP methods, eCLIP relies on RBP-RNA interactions covalently linked using UV crosslinking of live cells.
After ligation of a 3’ RNA adapter, immunoprecipitated material (as well as a paired input sample) are run on denaturing protein gels and transferred to nitrocellulose membranes.
eCLIP can also be used to identify miRNA targets and profile RNA modifications such as m6A.eCLIP datasets have been produced for over 150 RBPs with validated commercially available antibodies.
The method is based on linear amplification of the immunoprecipitated RNA and thereby improves the complexity of the sequencing-library despite significantly reducing the amount of input material and omitting several purification steps.
Along with a bioinformatical platform this method is designed to provide deep insights into RNA–protein interactomes in biomedical science, where the amount of starting material is often limited (i.e. in case of precious clinical samples).
As a modification of CLIP, methylated RNA sites were identified with the use of mutant enzyme or modification-specific antibody with the methods termed miCLIP or m6A-CLIP.