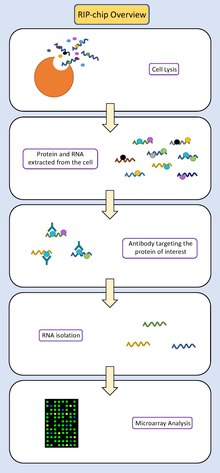
RNA immunoprecipitation chip
RIP-chip (RNA immunoprecipitation chip) is a molecular biology technique which combines RNA immunoprecipitation with a microarray.
[1][2][3][4] It can also be used to determine relative levels of gene expression, to identify subsets of RNAs which may be co-regulated, or to identify RNAs that may have related functions.
The strength of the fluorescent signal for a particular gene can indicate how much of that particular RNA was present in the original sample, which indicates the expression level of that gene.
Previous techniques aiming to understand protein-RNA interactions included RNA Electrophoretic Mobility Shift Assays and UV-crosslinking followed by RT-PCR,[9] however such selective analysis cannot be used when the bound RNAs are not yet known.
[1] To resolve this, RIP-chip combines RNA immunoprecipitation to isolate RNA molecules interacting with specific proteins with a microarray which can elucidate the identity of the RNAs participating in this interaction.