ELISA
ELISA has been used as a diagnostic tool in medicine, plant pathology, and biotechnology, as well as a quality control check in various industries.
Between each step, the plate is typically washed with a mild detergent solution to remove any proteins or antibodies that are non-specifically bound.
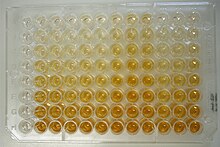
After the final wash step, the plate is developed by adding an enzymatic substrate to produce a visible signal, which indicates the quantity of antigen in the sample.
As an analytical biochemistry assay and a "wet lab" technique, ELISA involves detection of an analyte (i.e., the specific substance whose presence is being quantitatively or qualitatively analyzed) in a liquid sample by a method that continues to use liquid reagents during the analysis (i.e., controlled sequence of biochemical reactions that will generate a signal which can be easily quantified and interpreted as a measure of the amount of analyte in the sample) that stays liquid and remains inside a reaction chamber or well needed to keep the reactants contained.
Even if the sample is liquid (e.g., a measured small drop), the final detection step in "dry" analysis involves reading of a dried strip by methods such as reflectometry and does not need a reaction containment chamber to prevent spillover or mixing between samples.
The quantitative "reading" is usually based on detection of intensity of transmitted light by spectrophotometry, which involves quantitation of transmission of some specific wavelength of light through the liquid (as well as the transparent bottom of the well in the multiple-well plate format).
[7] Before the development of the ELISA, the only option for conducting an immunoassay was radioimmunoassay, a technique using radioactively labeled antigens or antibodies.
When enzymes (such as horseradish peroxidase) react with appropriate substrates (such as ABTS or TMB), a change in color occurs, which is used as a signal.
[10] In 1971, Peter Perlmann and Eva Engvall at Stockholm University in Sweden, and Anton Schuurs and Bauke van Weemen in the Netherlands independently published papers that synthesized this knowledge into methods to perform EIA/ELISA.
[11][12] Traditional ELISA typically involves chromogenic reporters and substrates that produce some kind of observable color change to indicate the presence of antigen or analyte.
Newer ELISA-like techniques use fluorogenic, electrochemiluminescent, and quantitative PCR reporters to create quantifiable signals.
[13][14] In technical terms, newer assays of this type are not strictly ELISAs, as they are not "enzyme-linked", but are instead linked to some nonenzymatic reporter.
In 2012, an ultrasensitive, enzyme-based ELISA test using nanoparticles as a chromogenic reporter was able to give a naked-eye colour signal, from the detection of mere attograms of analyte.
The sandwich or indirect ELISA provides a solution to this problem, by using a "capture" antibody specific for the test antigen to pull it out of the serum's molecular mixture.
Two or three times the standard deviation (error inherent in a test) is often used to distinguish positive from negative samples.
In quantitative ELISA, the optical density (OD) of the sample is compared to a standard curve, which is typically a serial dilution of a known-concentration solution of the target molecule.
However, the use of a secondary-antibody conjugate avoids the expensive process of creating enzyme-linked antibodies for every antigen one might want to detect.
Therefore, a sandwich ELISA used for research often needs validation, to reduce the risk of false positive results.
The steps for this ELISA are somewhat different from the first two examples: Unlabeled antibody is incubated in the presence of its antigen (sample).
In an ELISA, a person's serum is diluted 400 times and applied to a plate to which HIV antigens are attached.
However, eSimoa advances this concept by enabling enzymatic reaction measurements at the single-molecule level, which dramatically improves detection limits for various enzymes and biomolecules.
The enhanced sensitivity of eSimoa is crucial for early and accurate biomarker detection in clinical diagnostics, facilitating better disease monitoring and management.
In drug discovery, the ability to track subtle changes in enzymatic activity aids in the development of more effective pharmaceuticals by providing detailed insights into enzyme inhibition mechanisms.
Chi-An Cheng at National Taiwan University (NTU) has claimed that her team developed this innovative technology.
[35][36] However, this claim is contested by the existence of prior publications by David R. Walt's team at Harvard University, who published their work on eSimoa in 2020.