Gene Disease Database
[2][3] Experts in different areas of biology and bioinformatics have been trying to comprehend the molecular mechanisms of diseases to design preventive and therapeutic strategies for a long time.
For some illnesses, it has become apparent that it is the right amount of animosity is made for not enough to obtain an index of the disease-related genes but to uncover how disruptions of molecular grids in the cell give rise to disease phenotypes.
Recent developments in bioinformatics and laboratory genetics have made possible the better delineation of certain malformation and mental retardation syndromes, so that their mode of inheritance can be understood.
So one of the main concerns in biological and biomedical research is to recognise the underlying mechanisms behind this intricate genetic phenotypes.
The Comparative Toxicogenomics Database, helps to understand about the effects of environmental compounds on human health by integrating data from curated scientific literature to describe biochemical interactions with genes and proteins, and links between diseases and chemicals, and diseases and genes or proteins.
[11] CTD contains curated data defining cross-species chemical–gene/protein interactions and chemical– and gene–disease associations to illuminate molecular mechanisms underlying variable susceptibility and environmentally influenced diseases.
It contains a large amount of information about the biological function of proteins derived from the study literature, which can hint to a direct connection between gene-protein-disease.
[17] This explosion of information highlighted the need for a centralized database to efficiently and effectively collect, manage, and distribute a rat-centric view of this data to researchers around the world.
[19] OMIM is a comprehensive, authoritative compendium of human genes and genetic phenotypes that is freely available and updated daily.
The set of consequence terms, defined by the Sequence Ontology (SO) can be currently assigned to each combination of an allele and a transcript.
SIFT can be applied to naturally occurring nonsynonymous polymorphisms and laboratory-induced missense mutations, that will lead to build relationships in phenotype characteristics, proteomics and genomics.
GAD is primarily focused on archiving information on common complex human disease rather than rare Mendelian disorders as found in the OMIM.
It includes curated summary data extracted from published papers in peer reviewed journals on candidate gene and genome Wide Association Studies (GWAS).
It uses a machine learning based algorithm to extract semantic gene-disease relations from a textual source of interest.
[24] This sort of databases include Mendelian, compound and environmental diseases in an integrated gene-disease association archive and show that the concept of modularity applies for all of them They provide a functional analysis of diseases in case of important new biological insights, which might not be discovered when considering each of the gene-disease associations independently.
[1] Some of the most interesting cases using Gene-Disease Databases can be found in the following papers:[1][8] The completion of the human genome has changed the way the search for disease genes is performed.
Now, projects like the DisGeNET exemplify the efforts to systematically analyze all the gene alterations involved in a single or multiple diseases.
[1] Bioinformatics is both a term for the body of biological gene disease studies that use computer programming as part of their methodology, as well as a reference to specific analysis pipelines that are repeatedly used, particularly in the fields of genetics and genomics.
Often, such identification is made with the aim of better understanding the genetic basis of disease, unique adaptations, desirable properties, or differences between populations.
For instance, the availability of large numbers of individual human genomes will promote the development of computational analyses of rare variants, including the statistical mining of their relations to lifestyles, drug interactions and other factors.
In silico studies of the relationships between human variations and their effect on diseases will be key to the development of personalized medicine.
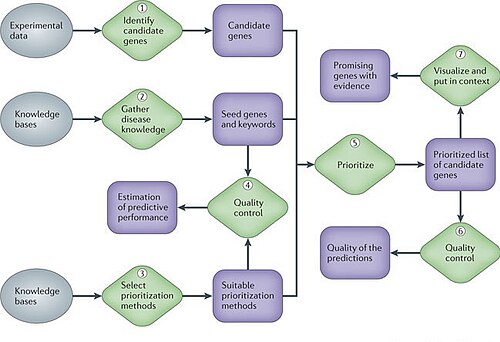

The curated data may comprise a process from practical experience and literature review to web publication of the database [ 14 ]


The description of each association type in this ontology is: #Therapeutic Association: The gene/protein has a therapeutic role in the amelioration of the disease. #Biomarker Association: The gene/protein either plays a role in the etiology of the disease (e.g. participates in the molecular mechanism that leads to disease) or is a biomarker for a disease. #Genetic Variation Association: Used when a sequence variation (a mutation, a SNP) is associated to the disease phenotype, but there is still no evidence to say that the variation causes the disease. In some cases the presence of the variants increase the susceptibility to the disease. In general, the NCBI SNP identifiers are provided. #Altered Expression Association: Alterations in the function of the protein by means of altered expression of the gene are associated with the disease phenotype. #Post-translational Modification Association: Alterations in the function of the protein by means of post-translational modifications (methylation or phosphorylation of the protein) are associated with the disease phenotype. [ 1 ]
