Haplogroup A (Y-DNA)
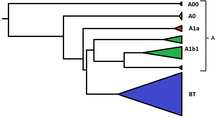
By this definition, haplogroup A includes all mutations that took place between the Y-chromosomal most recent common ancestor (estimated at some 270 kya) and the mutation defining haplogroup BT (estimated at some 140–150 kya),[8] including any extant subclades that may yet to be discovered.
[10] Scozzari et al. (2012) also supported "the hypothesis of an origin in the north-western quadrant of the African continent for the A1b [ i.e. A0 ] haplogroup".
[11] Haplogroup A1b1b2 has been found among ancient fossils excavated at Balito Bay in KwaZulu-Natal, South Africa, which have been dated to around 2149-1831 BP (2/2; 100%).
[12] By definition of haplogroup A as "non-BT", it is almost completely restricted to Africa, though a very small handful of bearers have been reported in Europe and Western Asia.
However, haplogroup A's oldest sub-clades are exclusively found in Central-Northwest Africa, where it (and by extension the patrilinear ancestor of modern humans) is believed to have originated.
[17][18] According to Wood et al. (2005) and Rosa et al. (2007), such relatively recent population movements from West Africa changed the pre-existing population Y chromosomal diversity in Central, Southern and Southeastern Africa, replacing the previous haplogroups in these areas with the now dominant E1b1a lineages.
Traces of ancestral inhabitants, however, can be observed today in these regions via the presence of the Y DNA haplogroups A-M91 and B-M60 that are common in certain relict populations, such as the Mbuti Pygmies and the Khoisan.
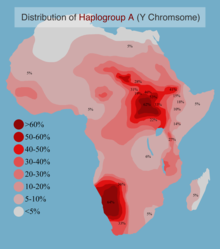
[29] The highest frequencies of haplogroup A have been reported among the Khoisan of Southern Africa, Beta Israel, and Nilo-Saharans from Sudan.
1 African American Male out of Lacrosse, WI USA, Moses, Ramon, A00, A00-AF8 In North Africa, haplogroup A is largely absent.
[23] Further downstream around the Nile valley, the subclade A3b2 has also been observed at very low frequencies in a sample of Egyptian males (3%).
[31] Haplogroup A3b2-M13 has been observed in populations of northern Cameroon (2/9 = 22% Tupuri,[20] 4/28 = 14% Mandara,[20] 2/17 = 12% Fulbe[24]) and eastern DRC (2/9 = 22% Alur,[20] 1/18 = 6% Hema,[20] 1/47 = 2% Mbuti[20]).
It has an estimated age of around 275 kya,[14][15] so is roughly contemporary with the known appearance of earliest known anatomically modern humans, such as Jebel Irhoud.
This previously unknown haplogroup was discovered in 2012 in the Y chromosome of an African-American man who had submitted his DNA for commercial genealogical analysis.
Further research in 2015 indicates that the modern population with the highest concentration of A00 is the Bangwa [fr] (or Nweh), a Yemba-speaking group of Cameroon (Grassfields Bantu): 27 of 67 (40.3%) samples were positive for A00a (L1149).
[40] Geneticists sequenced genome-wide DNA data from four people buried at the site of Shum Laka in Cameroon between 8000–3000 years ago, who were most genetically similar to Mbuti pygmies.
[11][failed verification] The subclade A1a (M31) has been found in approximately 2.8% (8/282) of a pool of seven samples of various ethnic groups in Guinea-Bissau, especially among the Papel-Manjaco-Mancanha (5/64 = 7.8%).
It was discovered that these men had a common male-line ancestor from the 18th century, but no previous information about African ancestry was known.
The subclade (appropriately considered as a distinct haplogroup) A1b1b1 (M28; formerly A3a) has only been rarely observed in the Horn of Africa.
[23] Haplogroup A-M13 also has been observed occasionally outside of Central and Eastern Africa, as in the Aegean Region of Turkey (2/30 = 6.7%[45]), Yemenite Jews (1/20 = 5%[25]), Egypt (4/147 = 2.7%,[27] 3/92 = 3.3%[20]), Palestinian Arabs (2/143 = 1.4%[46]), Sardinia (1/77 = 1.3%,[47] 1/22 = 4.5%[23]), the capital of Jordan, Amman (1/101=1%[48]), and Oman (1/121 = 0.8%[27]).
Haplogroup A-M13 has been found among three Neolithic period fossils excavated from the Kadruka site in Sudan.
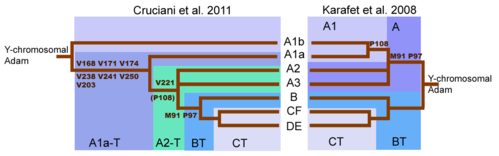
While the SNP marker M91 had been regarded as a key to identifying haplogroup BT, it was realised that the region surrounding M91 was a mutational hotspot, which is prone to recurrent back-mutations.