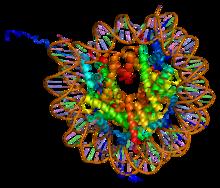
Histone H2A
One chromatin molecule is composed of at least one of each core histones per 100 base pairs of DNA.
[4] The term "Histone H2A" is intentionally non-specific and refers to a variety of closely related proteins that vary often by only a few amino acids.
This was observed in differentiating neurons during synthesis and turnover; changes in variant composition were seen among the H2A.1 histone.
[5] Physically, there are small changes on the surface area of the nucleosome that make the histone differ from H2A.
Recent research suggests that H2AZ is incorporated into the nucleosome using a Swr1, a Swi2/Snf2- related adenosine triphosphatase.
The method of repair this variant employs is non-homologous end joining.
Direct DNA damage can induce changes to the sequence variants.
Experiments performed with ionizing radiation linked γ- phosphorylation of H2AX to double-strand breaks.
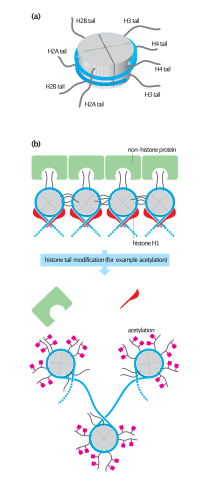
This variant differs from H2A because of the addition of a fold domain in its C-terminal tail.
The histone fold is a three-helix core domain that is connected by two loops.
This connection forms a ‘handshake arrangement.’ Most notably, this is termed the helix-turn-helix motif, which allows for dimerization with H2B.
The conserved domain contains a DNA binding structure and a peptidase fold.
[12] Recent studies also show that nucleosome assembly protein 1 is also used to transport of H2A into the nucleus so it can wrap DNA.
In addition, when H2A.Z was studied in human and yeast cells, it was used to promote RNA polymerase II recruitment.
In vertebrates and invertebrates, Histone H2A variant is reported to be involved in host immune response by acting as antimicrobial peptides (AMPs).
H2A are α-helical molecule, amphipathic protein with hydrophobic and hydrophilic residues on opposing sides that enhances the antimicrobial activity of H2A.
[14] Site specific ubiquitination of histone H2A has a role in the recruitment of DNA repair proteins to DNA double strand breaks which then may be repaired by either homologous recombination or non-homologous end joining.
Researchers studied eukaryotic evolutionary lineages of histone proteins and found diversification among the regulatory genes.
The greatest differences were observed in core histone gene cis-regulatory sequence motifs and associated protein factors.
At first researchers figured that these genes were redundant; however, when a mutant H2A.Z1 was created, it resulted in lethality during mammalian tests.