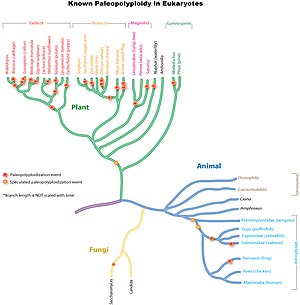
Paleopolyploidy
[8] Evidence suggests that baker's yeast (Saccharomyces cerevisiae), which has a compact genome, experienced polyploidization during its evolutionary history.
Studies suggest that the common ancestor of Poaceae, the grass family which includes important crop species such as maize, rice, wheat, and sugar cane, shared a whole genome duplication about 70 million years ago.
[10] In more ancient monocot lineages one or likely multiple rounds of additional whole genome duplications had occurred, which were however not shared with the ancestral eudicots.
[11] Further independent more recent whole genome duplications have occurred in the lineages leading to maize, sugar cane and wheat, but not rice, sorghum or foxtail millet.
[citation needed] A polyploidy event 160 million years ago is theorized to have created the ancestral line that led to all modern flowering plants.
[citation needed] A well-supported paleopolyploidy has been found in baker's yeast (Saccharomyces cerevisiae), despite its small, compact genome (~13Mbp), after the divergence from Kluyveromyces lactis and K.
[20] Duplication events that occurred a long time ago in the history of various evolutionary lineages can be difficult to detect because of subsequent diploidization (such that a polyploid starts to behave cytogenetically as a diploid over time) as mutations and gene translations gradually make one copy of each chromosome unlike its counterpart.
[clarification needed] Overall, paleopolyploidy can have both short-term and long-term evolutionary effects on an organism's fitness in the natural environment.
The duplicated genes can undergo neofunctionalization, subfunctionalization, or nonfunctionalization which could help the organism adapt to the new environment or survive different stress conditions.
[23] It has been suggested that many polyploidization events created new species, via a gain of adaptive traits, or by sexual incompatibility with their diploid counterparts.
An example would be the recent speciation of allopolyploid Spartina — S. anglica; the polyploid plant is so successful that it is listed as an invasive species in many regions.
This fact can be exploited in a laboratory setting by using colchicine to inhibit chromosome segregation during meiosis, creating synthetic autopolyploid plants.
[25][28] Polyploidy events will result in higher levels of heterozygosity, and, over time, can lead to an increase in the total number of functional genes in the genome.
[29] In one species, Glycine dolichocarpa (a close relative of the soybean, Glycine max), it has been observed that following a genome duplication roughly 500,000 years ago, there has been a 1.4 fold increase in transcription, indicating that there has been a proportional decrease in transcription relative to gene copy number following the duplication event.
[32] Additional arguments against 2R were based on the lack of the (AB)(CD) tree topology amongst four members of a gene family in vertebrates.