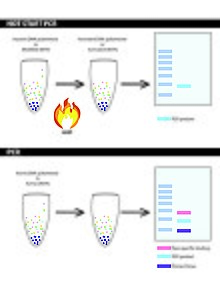
Hot start PCR
[5] Non-specific binding and priming or formation of primer dimers are minimized by completing the reaction mix after denaturation.
These modifications work overall to ensure that specific enzymes in solution will remain inactive or are inhibited until the optimal annealing temperature is reached.
Polymerase chain reaction (PCR) is a molecular biology technique used to amplify specific DNA segments by several orders of magnitude.
Similarly, primer dimers form complexes which decreases the amount of copy number amplifications obtained.
[10] This can be controlled by implementing hot start PCR which allows primer extensions to be blocked until the optimal temperatures are met.
[5][2] Hot start PCR can also occur when the Taq polymerase is inhibited/inactivated or its addition is delayed until optimal annealing temperatures, through deoxyribonucleotide triphosphate modifications or by modifying the primers through caging and secondary structure manipulation.
The antibodies link and bind to the polymerase, preventing early DNA amplification which could occur at lower temperatures.
Once the optimal annealing temperature is met, the antibodies will begin to degrade and dissociate, releasing the Taq DNA polymerase into the reaction and allowing the amplification process to start.
Once the temperature rises over 70 °C, during the denaturation step in the first cycle, the wax bead melts, allowing the Taq DNA polymerase to escape past the barrier and be released into the reaction – starting the amplification process.
Highly specific oligonucleotides, such as aptamers, bind to Taq DNA polymerase at lower temperatures making it inactive in the mixture.
[15] Another method is through deoxyribonucleotide triphosphate mediated hot start PCR which modifies the nucleotide bases through a protecting group.
Hot start dNTP can be chemically modified to include a heat sensitive protecting group at the 3 prime terminus.
This modification will prevent the nucleotides from interacting with the Taq polymerase to bind to the template strand until after the optimal temperatures are reached therefore, the protecting group will be removed during the heat activation step.
Similarly, hot start PCR inhibits the binding of primers to the template sequences which have a low homology which leads to mispriming.
It can also improve specificity and sensitivity, due to the stringent conditions, as well as increase the product yield of the targeted fragment.
[5] In antibody based hot start PCR, the polymerase is activated after the initial denaturation step during the cycling process, therefore decreasing the time required.