DNA polymerase
Before replication can take place, an enzyme called helicase unwinds the DNA molecule from its tightly woven form, in the process breaking the hydrogen bonds between the nucleotide bases.
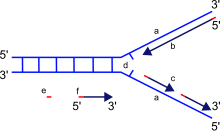
Since DNA polymerase requires a free 3' OH group for initiation of synthesis, it can synthesize in only one direction by extending the 3' end of the preexisting nucleotide chain.
The function of DNA polymerase is not quite perfect, with the enzyme making about one mistake for every billion base pairs copied.
Many DNA polymerases contain an exonuclease domain, which acts in detecting base pair mismatches and further performs in the removal of the incorrect nucleotide to be replaced by the correct one.
[10] The shape and the interactions accommodating the Watson and Crick base pair are what primarily contribute to the detection or error.
The loss of an interaction, which occurs at a mismatch, is said to trigger a shift in the balance, for the binding of the template-primer, from the polymerase, to the exonuclease domain.
Relative to the shape of DNA polymerase's binding pocket, steric clashes occur between the purine and residues in the minor groove, and important van der Waals and electrostatic interactions are lost by the pyrimidine.
In the case of DNA polymerase, the degree of processivity refers to the average number of nucleotides added each time the enzyme binds a template.
Instead, Pol I starts adding nucleotides at the RNA primer:template junction known as the origin of replication (ori).
Approximately 400 bp downstream from the origin, the Pol III holoenzyme is assembled and takes over replication at a highly processive speed and nature.
[24] Pfu DNA polymerase is a heat-stable enzyme of this family found in the hyperthermophilic archaeon Pyrococcus furiosus.
It consists of three assemblies: the pol III core, the beta sliding clamp processivity factor, and the clamp-loading complex.
The old textbook "trombone model" depicts an elongation complex with two equivalents of the core enzyme at each replication fork (RF), one for each strand, the lagging and leading.
[24] However, recent evidence from single-molecule studies indicates an average of three stoichiometric equivalents of core enzyme at each RF for both Pol III and its counterpart in B. subtilis, PolC.
[29] In-cell fluorescent microscopy has revealed that leading strand synthesis may not be completely continuous, and Pol III* (i.e., the holoenzyme α, ε, τ, δ and χ subunits without the ß2 sliding clamp) has a high frequency of dissociation from active RFs.
This suggests that the DnaB helicase may remain stably associated at RFs and serve as a nucleation point for the competent holoenzyme.
In vitro single-molecule studies have shown that Pol III* has a high rate of RF turnover when in excess, but remains stably associated with replication forks when concentration is limiting.
[30] Another single-molecule study showed that DnaB helicase activity and strand elongation can proceed with decoupled, stochastic kinetics.
[32] Another function of Pol IV is to perform translesion synthesis at the stalled replication fork like, for example, bypassing N2-deoxyguanine adducts at a faster rate than transversing undamaged DNA.
Cells lacking the dinB gene have a higher rate of mutagenesis caused by DNA damaging agents.
[18] DP1, a Mre11-like exonuclease,[39] is likely the precursor of small subunit of Pol α and ε, providing proofreading capabilities now lost in Eukaryotes.
[26] Its N-terminal HSH domain is similar to AAA proteins, especially Pol III subunit δ and RuvB, in structure.
TdT is expressed only in lymphoid tissue, and adds "n nucleotides" to double-strand breaks formed during V(D)J recombination to promote immunological diversity.
[26] Pol ε is unique in that it has two zinc finger domains and an inactive copy of another family B polymerase in its C-terminal.
The presence of this zinc finger has implications in the origins of Eukaryota, which in this case is placed into the Asgard group with archaeal B3 polymerase.
Pol η is particularly important for allowing accurate translesion synthesis of DNA damage resulting from ultraviolet radiation.
The gradual decrease in size of telomeres as the result of many replications over a lifetime are thought to be associated with the effects of aging.
[54][55] Any mutation that leads to limited or non-functioning Pol γ has a significant effect on mtDNA and is the most common cause of autosomal inherited mitochondrial disorders.
Pol θ extends mismatched primer termini and can bypass abasic sites by adding a nucleotide.
[67] It was proposed that a mutational alteration in the phage DNA polymerase can stimulate template strand switching (copy choice recombination) during replication.