COLD-PCR
Detection of mutations is important in the case of early cancer detection from tissue biopsies and body fluids such as blood plasma or serum, assessment of residual disease after surgery or chemotherapy, disease staging and molecular profiling for prognosis or tailoring therapy to individual patients, and monitoring of therapy outcome and cancer remission or relapse.
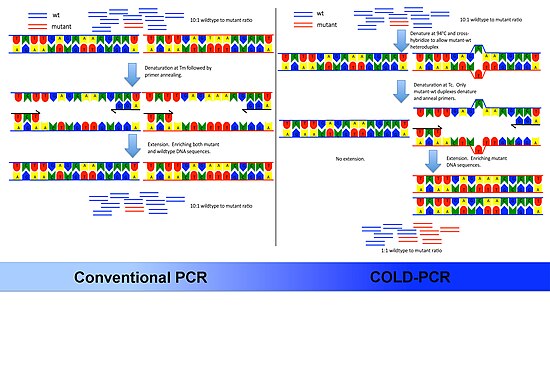
Common PCR will amplify both the major (wildtype) and minor (mutant) alleles with the same efficiency, occluding the ability to easily detect the presence of low-level mutations.
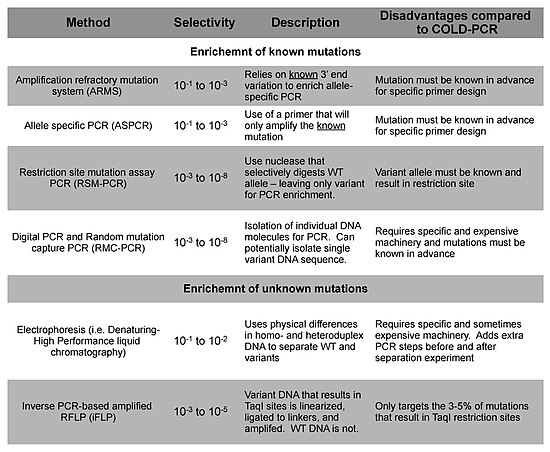
Replacing traditional PCR with COLD-PCR for these downstream assays will increase the reliability in detecting mutations from mixed samples, including tumors and body fluids.
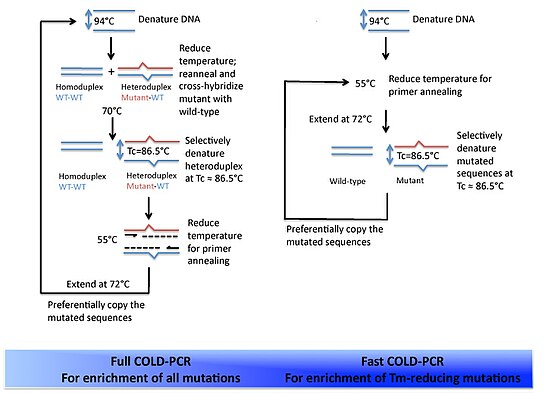
The underlying principle of COLD-PCR is that single nucleotide mismatches will slightly alter the melting temperature (Tm) of the double-stranded DNA.
[1] Comparison of Sanger sequencing chromatograms indicated that the mutant allele was enriched 13 fold when COLD-PCR was used compared to traditional PCR alone.
The study was able to detect 8 low level (under 20% abundance) mutations that would likely have been missed using conventional methods that don't enrich for variant sequence DNA.
The same research group that developed COLD-PCR and used it to compare the sensitivity of regular PCR for genotyping with direct Sanger sequencing, RFLP, and pyrosequencing, also ran a similar study using MALDI-TOF as a downstream application for detecting mutations.
COLD-PCR run on a quantitative PCR machine, using TaqMan probes specific for a mutation, was shown to increase the measured difference between mutant and wildtype samples.
[4] COLD-PCR was originally described by Li et al. in a Nature Medicine paper published in 2008 from Mike Makrigiorgos's lab group at the Dana Farber Cancer Institute of Harvard Medical School.